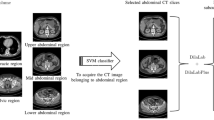
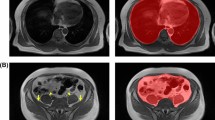
Abstract
Purpose
Fully automated abdominal adipose tissue segmentation from computed tomography (CT) scans plays an important role in biomedical diagnoses and prognoses. However, to identify and segment subcutaneous adipose tissue (SAT) and visceral adipose tissue (VAT) in the abdominal region, the traditional routine process used in clinical practise is unattractive, expensive, time-consuming and leads to false segmentation. To address this challenge, this paper introduces and develops an effective global-anatomy-level convolutional neural network (ConvNet) automated segmentation of abdominal adipose tissue from CT scans termed EFNet to accommodate multistage semantic segmentation and high similarity intensity characteristics of the two classes (VAT and SAT) in the abdominal region.
Methods
EFNet consists of three pathways: (1) The first pathway is the max unpooling operator, which was used to reduce computational consumption. (2) The second pathway is concatenation, which was applied to recover the shape segmentation results. (3) The third pathway is anatomy pyramid pooling, which was adopted to obtain fine-grained features. The usable anatomical information was encoded in the output of EFNet and allowed for the control of the density of the fine-grained features.
Results
We formulated an end-to-end manner for the learning process of EFNet, where the representation features can be jointly learned through a mixed feature fusion layer. We immensely evaluated our model on different datasets and compared it to existing deep learning networks. Our proposed model called EFNet outperformed other state-of-the-art models on the segmentation results and demonstrated tremendous performances for abdominal adipose tissue segmentation.
Conclusion
EFNet is extremely fast with remarkable performance for fully automated segmentation of the VAT and SAT in abdominal adipose tissue from CT scans. The proposed method demonstrates a strength ability for automated detection and segmentation of abdominal adipose tissue in clinical practise.





Similar content being viewed by others
References
Fu Y, Ippolito JE, Ludwig DR, Nizamuddin R, Li HH, Yang D (2020) Automatic segmentation of ct images for ventral body composition analysis. Med Phys 47(11):5723–5730
Aljunid SM (2021) Obesity, a costly epidemic. Laparoscopic sleeve gastrectomy. Springer, Berlin, pp 13–22
Kissebah AH, Vydelingum N, Murray R, Evans DJ, Hartz AJ, Kalkhoff RK, Adams PW (1982) Relation of body fat distribution to metabolic complications of obesity. J Clin Endocrinol Metab 54(2):254–260
Colditz GA, Peterson LL (2018) Obesity and cancer: evidence, impact, and future directions. Clin Chem 64(1):154–162
Napolitano A, Miller SR, Murgatroyd PR, Coward WA, Wright A, Finer N, De Bruin TW, Bullmore ET, Nunez DJ (2008) Validation of a quantitative magnetic resonance method for measuring human body composition. Obesity 16(1):191–198
Havaei M, Davy A, Warde-Farley D, Biard A, Courville A, Bengio Y, Pal C, Jodoin PM, Larochelle H (2017) Brain tumor segmentation with Deep Neural Networks. Med Image Anal 35:18–31
Dubrovina A, Kisilev P, Ginsburg B, Hashoul S, Kimmel R (2018) Computational mammography using deep neural networks. Comput Methods Biomech Biomed Eng: Imaging Visual 6(3):243–247
Kullberg J, Hedström A, Brandberg J, Strand R, Johansson L, Bergström G, Ahlström H (2017) Automated analysis of liver fat, muscle and adipose tissue distribution from CT suitable for large-scale studies. Scientif Rep 7(1):1–11
Wang Y, Qiu Y, Thai T, Moore K, Hong L, Zheng B (2017) A two-step convolutional neural network based computer-aided detection scheme for automatically segmenting adipose tissue volume depicting on CT images. Comput Methods Programs Biomed 144:97–104
Langner T, Hedstrom A, Morwald K, Weghuber D, Forslund A, Bergsten P, Ahlstrom H, Kullberg J (2019) Fully convolutional networks for automated segmentation of abdominal adipose tissue depots in multicenter water–fat MRI. Magn Reson Med 81(4):2736–2745
Anthimopoulos MM, Christodoulidis S, Ebner L, Geiser T, Christe A, Mougiakakou SG (2018) Semantic segmentation of pathological lung tissue with dilated fully convolutional networks. IEEE J Biomed Health Informatics 23(2):714–722
Badrinarayanan V, Kendall A, Cipolla R (2017) Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Machine Intell 39(12):2481–2495
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. In: International Conference on Medical Image Computing & Computer-assisted Intervention.
Zhao H, Shi J, Qi X, Wang X, Jia J (2017) Pyramid scene parsing network In: The IEEE Conference on Computer Vision and Pattern Recognition (CVPR)
Turchenko V, Chalmers E, Luczak A (2017) A deep convolutional auto-encoder with pooling - unpooling layers in caffe. Int J Comput 18:1
Estrada S, Lu R, Conjeti S, Orozco-Ruiz X, Panos-Willuhn J, Breteler M, Reuter M (2020) Fatsegnet: A fully automated deep learning pipeline for adipose tissue segmentation on abdominal dixon mri. Magn Reson Med 84(3):1471–1483
Weston AD, Korfiatis P, Kline TL, Philbrick KA, Kostandy P, Sakinis T, Sugimoto M, Takahashi N, Erickson BJ (2019) Automated abdominal segmentation of ct scans for body composition analysis using deep learning. Radiology 290(3):669–679
Agarwal C, Dallal AH, Arbabshirani MR, Patel A, Moore G (2017) Unsupervised quantification of abdominal fat from CT images using greedy snakes. In: Society of Photo-Optical Instrumentation Engineers, p 101332T
Hussein BR, Malik OA, Ong W-H, Slik JWF (2020) Semantic segmentation of herbarium specimens using deep learning techniques, lecture notes in electrical. Engineering 603:321–330
WickstrØm K, Kampffmeyer M, Jenssen R (2020) Uncertainty and interpretability in convolutional neural networks for semantic segmentation of colorectal polyps. Med Image Anal 60:101619
Xia H, Sun W, Song S, Mou X (2020) Md-net: multi-scale dilated convolution network for ct images segmentation. Neural Process Lett 51(3):2915–2927
Chen LC, Papandreou G, Kokkinos I, Murphy K, Yuille AL (2018) DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans Pattern Anal Mach Intell 40(4):834–848
Roy AG, Conjeti S, Navab N, Wachinger C (2019) Quicknat: A fully convolutional network for quick and accurate segmentation of neuroan atomy. Neuroimage 186:713–727
https://www.tensorflow.org/api_docs/python/tf/nn/max_pool_with_argmax.
Goroshin R, Mathieu M, LeCun Y (2015) Learning to linearize under uncertainty. In: Advances in Neural Information Processing Systems, pp 1234–1242
Zhao J, Mathieu M, Goroshin R, Lecun Y (2016) Stacked What-Where Auto-encoders. Comput Sci 15(1):3563–3593
Pinheiro PHO, Collobert R (2014) Recurrent convolutional neural networks for scene labeling. In: The 31st International Conference on Machine Learning, Vol. 32
Zhou B, Khosla A, Lapedriza A, Oliva A, Torralb A (2014) Object detectors emerge in deep scene CNNs. In: Computer Science
Chen LC, Zhu Y, Papandreou G, Schroff F, Adam H (2018) Encoder-decoder with atrous separable convolution for semantic image segmentation. In: The European Conference on Computer Vision (ECCV)
Manaswi NK (2018) Understanding and working with keras, In: Deep Learning with Applications Using Python: Chatbots and Face, Object, and Speech Recognition With TensorFlow and Keras, pp 31–43
Girija SS (2016) Tensorflow: Large-scale machine learning on heterogeneous distributed systems, Software available from tensorflow.org 39(9)
Jia S, Jiang S, Lin Z, Li N, Xu M, Yu S (2021) A survey: Deep learning for hyperspectral image classification with few labeled samples. Neurocomputing 448:179–204
Yan K, Wang X, Lu L, Summers RM (2018) Deeplesion: automated mining of large-scale lesion annotations and universal lesion detection with deep learning. J Med Imaging 5(3):036501
Fatima N (2020) Enhancing performance of a deep neural network: A comparative analysis of optimization algorithms. ADCAIJ Adv Distributed Comput Artif Intell J 9(2):79–90
Hinton G, Tieleman T (2012) Lecture 6.5-rmsprop: divide the gradient by a running average of its recent magnitude. COURSERA Neural Netw Mach Learn 4:26–30
Klein S, Van-der Heide UA, Lips IM, Van-Vulpen M, Staring M, Pluim JP (2008) Automatic segmentation of the prostate in 3D MR images by atlas matching using localized mutual information. Med Phys 35(4):1407–1417
Dosovitskiy A, Fischer P, Springenberg JT, Riedmiller M, Brox T (2016) Discriminative unsupervised feature learning with exemplar convolutional neural networks. IEEE Trans Pattern Anal Mach Intell 38(9):1734–1747
Lazebnik S, Schmid C, Ponce J (2006) Beyond bags of features: spatial pyramid matching for recognizing natural scene categories. In: International Conference on Medical Image Computing & Computer-assisted Intervention, Vol. 2, pp 2169–2178
Wang Z, Meng Y, Weng F, Chen Y, Lu F, Liu X, Zhang J (2020) An effective cnn method for fully automated segmenting subcutaneous and visceral adipose tissue on ct scans. Annal Biomed Eng 48(1):312–328
Funding
This work was supported by Scientific Research Fund of Hunan Provincial Education Department(grant number 20C0402) and by Hunan First Normal University(grant number XYS16N03), also funded by the Projects of the National Social Science Foundation of China (grant number 82073019 and 82073018) and Hunan National Applied Mathematics Centre (grant number 2020ZYT003).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, Z., Hounye, A.H., Zhang, J. et al. Deep learning for abdominal adipose tissue segmentation with few labelled samples. Int J CARS 17, 579–587 (2022). https://doi.org/10.1007/s11548-021-02533-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-021-02533-8