Abstract
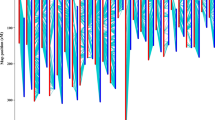
Quantitative trait locus (QTL) mapping is an important method in marker-assisted selection breeding. Many studies on the QTLs focus on cotton fibre yield and quality; however, most are conducted at the DNA level, which may reveal null QTLs. Hence, QTL mapping based on transcriptome maps at the cDNA level is often more reliable. In this study, an interspecific transcriptome map of allotetraploid cotton was developed based on an F2 population (Emian22 × 3-79) by amplifying cDNA using EST-SSRs. The map was constructed using cDNA obtained from developing fibres at five days post anthesis (DPA). A total of 1270 EST-SSRs were screened for polymorphisms between the mapping parents. The resulting transcriptome linkage map contained 242 markers that were distributed in 32 linkage groups (26 chromosomes). The full length of this map is 1938.72 cM with a mean marker distance of 8.01 cM. The functions of some ESTs have been annotated by exploring homologous sequences. Some markers were related to the differentiation and elongation of cotton fibre, while most were related to the basic metabolism. This study demonstrates that constructing a transcriptome linkage map by amplifying cDNAs using EST-SSRs is a simple and practical method as well as a powerful tool to map eQTLs for fibre quality and other traits in cotton.




Similar content being viewed by others
References
Brugmans B., Fernandez del Carmen A., Bachem C. W., van Os H., van Eck H. J., and Visser R. G. 2002 A novel method for the construction of genomewide transcriptome maps. Plant J. 31, 211–222.
Claverie M., Souquet M., Jean J., Forestier-Chiron N., Lepitre V., Pré M., et al. 2012 cDNA-AFLP-based genetical genomics in cotton fibers. Theor. Appl. Genet. 124, 665–683.
Conesa A., Götz S., Garcia-Gomez J. M., Terol J., Talon M., and Robles M. 2005 Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676.
Fang D. D. and Yu J. Z. 2012 Addition of 455 microsatellite marker loci to the high-density Gossypium hirsutum TM-1 × G. barbadense 3-79 genetic map. J. Cotton Sci. 16, 229–248.
Götz S., García-Gómez J. M., Terol J., Williams T. D., Nueda M. J., Robles M., et al. 2008 High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435.
He D. H., Lin Z. X., Zhang X. L., Nie Y. C., Guo X. P., Zhang Y. X., and Li W. 2007 QTL mapping for economic traits based on a dense genetic map of cotton with PCR-based markers using the interspecific cross of Gossypium hirsutum × Gossypium barbadense. Euphytica 153, 181–197.
He X. H., Qin H., Hu Z., Zhang T., and Zhang Y. M. 2011 Mapping of epistatic quantitative trait loci in four-way crosses. Theor. Appl. Genet. 122, 33–48.
Kosambi D. D. 1944 The estimation of map distance from recombination values. Ann. Eugen. 12, 172–175.
Lacape J. M., Nguyen T. B., Courtois B., Belot J. L., Giband M., Gourlot J. P., et al. 2005 QTL analysis of cotton fiber quality using multiple Gossypium hirsutum × Gossypium barbadense backcross generations. Crop Sci. 45, 123–140.
Lacape J. M., Llewellyn D., Jacobs J., Arioli T., Becker D., Calhoun S., et al. 2010 Meta-analysis of cotton fiber quality QTLs across diverse environments in a Gossypium hirsutum × G. barbadense RIL population. BMC Plant Biol. 10, 132.
Lee J. J., Woodward A. W., and Chen Z. J. 2007 Gene expression changes and early events in cotton fibre development. Ann. Bot. 100, 1391–1401.
Li G., Gao M., Yang B., and Quiros C. F. 2003 Gene for gene alignment between the Brassica and Arabidopsis genomes by direct transcriptome mapping. Theor. Appl. Genet. 107, 168–180.
Lin Z., He D., Zhang X., Nie Y., Guo X., Feng C., et al. 2005 Linkage map construction and mapping QTL for cotton fiber quality using SRAP, SSR and RAPD. Plant Breed. 124, 180–187.
Lin Z. X., Zhang Y. X., Zhang X. L., and Guo X. P. 2009 A high-density integrative linkage map for Gossypium hirsutum. Euphytica 166, 35–45.
Liu H. W., Wang X. F., Pan Y. X., Shi R. F., Zhang G. Y., and Ma Z. Y. 2009 Mining cotton fiber strength candidate genes based on transcriptome mapping. Chin. Sci. Bull. 54, 4651–4657.
Liu R., Wang B., Guo W., Wang L. and Zhang T. 2011 Differential gene expression and associated QTL mapping for cotton yield based on a cDNA-AFLP transcriptome map in an immortalized F2. Theor. Appl. Genet. 123, 439–454.
Liu R., Wang B., Guo W., Qin Y., Wang L., Zhang Y., and Zhang T. 2012 Quantitative trait loci mapping for yield and its components by using two immortalized populations of a heterotic hybrid in Gossypium hirsutum L. Mol. Breed. 29, 297–311.
Liu C., Yuan D., Zhang X., and Lin Z. 2013 Isolation, characterization and mapping of genes differentially expressed during fibre development between Gossypium hirsutum and G. barbadense by cDNA-SRAP. J. Genet. 92, 175–181.
Mei M., Syed N. H., Gao W., Thaxton P. M., Smith C. W., Stelly D. M., and Chen Z. J. 2004 Genetic mapping and QTL analysis of fiber-related traits in cotton (Gossypium). Theor. Appl. Genet. 108, 280–291.
Pang M., Percy R. G., Stewart J. Mc. D., Hughs E., and Zhang J. 2012 Comparative transcriptome analysis of Pima and Acala cotton during boll development using 454 pyrosequencing technology. Mol. Breed. 30, 1143–1153.
Qin H. D., Guo W. Z., Zhang Y. M., and Zhang T. Z. 2008 QTL mapping of yield and fiber traits based on a four-way cross population in Gossypium hirsutum L. Theor. Appl. Genet. 117, 883– 894.
Reinisch A. J., Dong J. M., Brubaker C. L., Stelly D. M., Wendel J. F., and Paterson A. H. 1994 A detailed RFLP map of cotton Gossypium hirsutum × Gossypium barbadense: chromosome organization and evolution in a disomic polyploid genome. Genetics 138, 829–847.
Ritter E., de Galarreta J. I. R., van Eck H. J., and Sánchez I. 2008 Construction of a potato transcriptome map based on the cDNA-AFLP technique. Theor. Appl. Genet. 116, 1003–1013.
Rong J. K., Abbey C., Bowers J. E., Brubaker C. L., Chang C., Peng W. C., et al. 2004 A 3347-locus genetic recombination map of sequence-tagged sites reveals features of genome organization, transmission and evolution of cotton (Gossypium). Genetics 166, 389–417.
Shen X. L., Zhang T. Z., Guo W. Z., Zhu X. F., and Zhang X. Y. 2006 Mapping fiber and yield QTLs with main, epistatic, and QTL × environment interaction effects in recombinant inbred lines of upland cotton. Crop Sci. 46, 61–66.
Stam P. 1993 Construction of integrated genetic linkage maps by means of a new computer package: join map. Plant J. 3, 739–744.
Sun F. D., Zhang J. H., Wang S. F., Gong W. K., Shi Y. Z., Liu A. Y., et al. 2012 QTL mapping for fiber quality traits across multiple generations and environments in upland cotton. Mol. Breed. 30, 569–582.
Ulloa M., Saha S., Jenkins J. N., Meredith W. R., McCarty J. C., and Stelly D. M. 2005 Chromosomal assignment of RFLP linkage groups harboring important QTLs on an intraspecific cotton (Gossypium hirsutum L.) joinmap. J. Hered. 96, 132–144.
Wang B. H., Guo W. Z., Zhu X. F., Wu Y. T., Huang N. T., and Zhang T. Z. 2006 QTL mapping of fiber quality in an elite hybrid derived-RIL population of upland cotton. Euphytica 152, 367–378.
Wu J., Gutierrez O. A., Jenkins J. N., McCarty J. C., and Zhu J. 2009 Quantitative analysis and QTL mapping for agronomic and fiber traits in an RI population of upland cotton. Euphytica 165, 231–245.
Yu Y., Yuan D. J., Liang S. G., Li X. M., Wang X. Q., Lin Z. X., et al. 2011 Genome structure of cotton revealed by a genome-wide SSR genetic map constructed from a BC1 population between Gossypium hirsutum and G. barbadense. BMC Genomics 12, 15.
Yu J., Zhang K., Li S., Yu S., Zhai H., Wu M., et al. 2013 Mapping quantitative trait loci for lint yield and fiber quality across environments in a Gossypium hirsutum × Gossypium barbadense backcross inbred line population. Theor. Appl. Genet. 126, 275–287.
Zhang Z. S., Xiao Y. H., Luo M., Li X. B., Luo X. Y., Hou L., et al. 2005 Construction of a genetic linkage map and QTL analysis of fiber-related traits in upland cotton (Gossypium hirsutum L.) Euphytica 144, 91–99.
Zhang Z. S., Hu M. C., Zhang J., Liu D. J., Zheng J., Zhang K., et al. 2009 Construction of a comprehensive PCR-based marker linkage map and QTL mapping for fiber quality traits in upland cotton (Gossypium hirsutum L). Mol. Breed. 24, 49–61.
Zhang K., Zhang J., Ma J., Tang S., Liu D., Teng Z., et al. 2012 Genetic mapping and quantitative trait locus analysis of fiber quality traits using a three-parent composite population in upland cotton (Gossypium hirsutum L.) Mol. Breed. 29, 335–348.
Zhao L., Lv Y., Cai C., Tong X., Chen X., Zhang W., et al. 2012 Toward allotetraploid cotton genome assembly: integration of a high-density molecular genetic linkage map with DNA sequence information. BMC Genomics 13, 539.
Zhu L. F., Tu L. L., Zhen F. C., Liu D. Q., and Zhang X. L. 2005 An improved simple protocol for isolation of high quality RNA from Gossypium spp. suitable for cDNA library construction. Acta Agromomica Sin. 31, 1657–1659.
Acknowledgement
This work was financially supported by the National Basic Research Program (no. 2011CB109303 and 2010CB126001).
Author information
Authors and Affiliations
Corresponding author
Additional information
[Liu C., Yuan D. and Lin Z. 2014 Construction of an EST-SSR-based interspecific transcriptome linkage map of fibre development in cotton. J. Genet. 93, xx–xx]
Rights and permissions
About this article
Cite this article
LIU, C., YUAN, D. & LIN, Z. Construction of an EST-SSR-based interspecific transcriptome linkage map of fibre development in cotton. J Genet 93, 689–697 (2014). https://doi.org/10.1007/s12041-014-0425-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-014-0425-5