Abstract
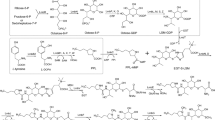
Amycolatopsis mediterranei S699 produces rifamycin B and successors of this strain are in use for the industrial production of rifamycin B. Semisynthetic derivatives of rifamycin B are used against Mycobacterium tuberculosis that causes tuberculosis. Although the rifamycin biosynthetic gene cluster was characterized two decades ago, the regulation of rifamycin B biosynthesis in Amycolatopsis mediterranei S699 is poorly understood. In this study, we analysed the genome and proteome of Amycolatopsis mediterranei S699 and identified 1102 transcription factors which comprise about 10% of the total genome. Using interactomics approaches we delineated 30 unique transcription factors directly involved in secondary metabolism that regulate rifamycin B biosynthesis. We also predict the role of RifN as hub in controlling the regulation of other genes involved in rifamycin biosynthesis. RifN is important for maintaining the integrity of the rifamycin-network. Thus, these transcription factor can be exploited to improve rifamycin B production in Amycolatopsis mediterranei S699.



Similar content being viewed by others
References
Romero-Rodríguez A, Robledo-Casados I, Sánchez S (2015) An overview on transcriptional regulators in Streptomyces. Biochim Biophys Acta 1849:1017–1039. https://doi.org/10.1016/j.bbagrm.2015.06.007
Seshasayee AS, Sivaraman K, Luscombe NM (2011) An overview of prokaryotic transcription factors: a summary of function and occurrence in bacterial genomes. Subcell Biochem 52:7–23. https://doi.org/10.1007/978-90-481-9069-0_2
Lal R, Kumari R, Kaur H, Khanna R, Dhingra N, Tuteja D (2000) Regulation and manipulation of the gene clusters encoding type –I PKSs. Trends Biotechnol 18:264–274. https://doi.org/10.1016/s0167-7799(00)01443-8
Aristoff PA, Garcia GA, Kirchhoff PD, Showalter HH (2010) Rifamycins–obstacles and opportunities. Tuberculosis 90:94–118. https://doi.org/10.1016/j.tube.2010.02.001
Kumari R, Singh P, Lal R (2016) Genetics and Genomics of the Genus Amycolatopsis. Indian J Microbiol 56:233–246. https://doi.org/10.1007/s12088-016-0590-8
Gatta L, Scarpignato C (2017) Systematic review with meta-analysis: rifaximin is effective and safe for the treatment of small intestine bacterial overgrowth. Aliment Pharmacol Ther 45:604–616. https://doi.org/10.1111/apt.13928
Kumar A, Chettiar S, Parish T (2017) Current challenges in drug discovery for tuberculosis. Expert Opin Drug Discov 12:1–4. https://doi.org/10.1080/17460441.2017.1255604
Schön T, Chryssanthou E (2017) Minimal inhibitory concentration distributions for Mycobacterium avium complex—towards evidence based susceptibility breakpoints. Int J Infect Dis 55:122–124. https://doi.org/10.1016/j.ijid.2016.12.027
Prasad R, Singh A, Balasubramanian V, Gupta N (2017) Extensively drug-resistant tuberculosis in India: current evidence on diagnosis & management. Indian J Med Res 145:271–293. https://doi.org/10.4103/ijmr.IJMR_177_16
Matteelli A, Roggi A, Carvalho AC (2014) Extensively drug-resistant tuberculosis: epidemiology and management. Clin Epidemiol 6:111–118. https://doi.org/10.2147/CLEPS35839
Haydel SE (2010) Extensively drug-resistant tuberculosis: a sign of the times and an impetus for antimicrobial discovery. Pharmaceuticals (Basel) 3:2268–2290. https://doi.org/10.3390/ph3072268
Shah NS, Wright A, Bai GH, Barrera L, Boulahbal F, Martín-Casabona N, Drobniewski F, Gilpin C, Havelková M, Lepe R, Lumb R, Metchock B, Portaels F, Rodrigues MF, Rüsch-Gerdes S, Van Deun A, Vincent V, Laserson K, Wells C, Cegielski JP (2007) Worldwide emergence of extensively drug-resistant tuberculosis. Emerg Infect Dis 13:380–387. https://doi.org/10.3201/eid1303.061400
August PR, Tang L, Yoon YJ, Ning S, Muller R, Yu TW, Taylor M, Hoffman D, Kim CG, Zhang X, Hutchinson CR, Floss HG (1998) Biosynthesis of the ansamycin antibiotic rifamycin: deductions from the molecular analysis of the rif biosynthetic gene cluster of Amycolatopsis mediterranei S699. Chem Biol 5:69–79. https://doi.org/10.1016/S1074-5521(98)90141-7
Xu J, Wan E, Kim CJ, Floss HG, Mahmud T (2005) Identification of tailoring genes involved in the modification of the polyketide backbone of rifamycin B by Amycolatopsis mediterranei S699. Microbiol 151:2515–2528. https://doi.org/10.1099/mic.0.28138-0
Nigam A, Almabruk KH, Saxena A, Jongtae Y, Mukherjee U, Kaur H, Kohli P, Kumari R, Singh P, Zakharov LN, Singh Y, Mahmud T, Lal R (2014) Modification of rifamycin polyketide backbone in Amycolatopsis mediterranei leads to the production of a new rifamycin B analog with an improved drug activity against rifampicin-resistant Mycobacterium tuberculosis strains. J Biol Chem 289:21142–21152. https://doi.org/10.1074/jbc.M114.572636
Singh P, Kumari R, Lal R (2017) Bedaquiline: fallible hope against drug resistant tuberculosis. Indian J Microbiol 57:371–377. https://doi.org/10.1007/s12088-017-0674-0
McArthur M, Bibb MJ (2008) Manipulating and understanding antibiotic production in Streptomyces coelicolor A3(2) with decoy oligonucleotides. Proc Natl Acad Sci USA 105:1020–1025. https://doi.org/10.1073/pnas0710724105
Wang J, Xu J, Luo S, Ma Z, Bechthold A, Yu X (2018) AdpAsd, a positive regulator for morphological development and toyocamycin biosynthesis in Streptomyces diastatochromogenes 1628. Curr Microbiol 75:1345–1351. https://doi.org/10.1007/s00284-018-1529-6
Xu X, Wang J, Bechthold A, Ma Z, Yu X (2017) Selection of an efficient promoter and its application in toyocamycin production improvement in Streptomyces diastatochromogenes 1628. World J Microbiol Biotechnol 33:30–38. https://doi.org/10.1007/s11274-016-2194-1
Guo J, Frost JW (2002) Biosynthesis of 1-deoxy-1-imino-D-erythrose 4-phosphate: a defining metabolite in the aminoshikimate pathway. J Am Chem Soc 124:528–529. https://doi.org/10.1021/ja016963v
Verma M, Kaur J, Kumar M, Kumari K, Saxena A, Anand S, Nigam A, Ravi V, Raghuvanshi S, Khurana P, Tyagi AK, Khurana JP, Lal R (2011) Whole genome sequence of Rifamycin B- producing strain Amycolatopsis mediterranei S699. J Bacteriol 193:5562–5563. https://doi.org/10.1128/JB.05819-11
UniProt Consortium (2007) The universal protein resource (UniProt). Nucleic Acids Res 36:D190–195. https://doi.org/10.1093/nar/gkm895(Database issue)
Mering CV, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B (2003) STRING: a database of predicted functional associations between proteins. Nucleic Acids Res 31:258–261. https://doi.org/10.1093/nar/gkg034
Ortet P, De Luca G, Whitworth DE, Barakat M (2012) P2TF: a comprehensive resource for analysis of prokaryotic transcription factors. BMC Genomics 13:628–636. https://doi.org/10.1186/1471-2164-13-628
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
Gupta V, Haider S, Sood U, Gilbert JA, Ramjee M, Forbes K, Singh Y, Lopes BS, Lal R (2016) Comparative genomic analysis of novel Acinetobacter symbionts: a combined systems biology and genomics approach. Sci Rep 6:29043–29055. https://doi.org/10.1038/srep29043
Grant JR, Stothard P (2008) The CGView server: a comparative genomics tool for circular genomes. Nucleic Acids Res 36:W181–W184. https://doi.org/10.1093/nar/gkn179
Lei C, Wang J, Liu Y, Liu X, Zhao G, Wang J (2017) A feedback regulatory model for RifQ-mediated repression of rifamycin export in Amycolatopsis mediterranei. Microb Cell Fact 17:14–23. https://doi.org/10.1186/s12934-018-0863-5
Li C, Liu X, Lei C, Yan H, Shao Z, Wang Y, Zhao G, Wang J, Ding X (2017) RifZ (AMED_0655) is a pathway-specific regulator for rifamycin biosynthesis in Amycolatopsis mediterranei. Appl Environ Microbiol 83:e03201–e3216. https://doi.org/10.1128/AEM.03201-16
Świątek MA, Gubbens J, Bucca G, Song E, Yang YH, Laing E, Kim BG, Smith CP, van Wezel GP (2013) The ROK family regulator Rok7B7 pleiotropically affects xylose utilization, carbon catabolite repression, and antibiotic production in Streptomyces coelicolor. J Bacteriol 195:1236–1248. https://doi.org/10.1128/JB02191-12
Grove A (2013) MarR family transcription factors. Curr Biol 23:R142–143. https://doi.org/10.1016/jcub201301013
Verma H, Dhingra GG, Sharma M, Gupta V, Negi RK, Singh Y, Lal R (2020) Comparative genomics of Sphingopyxis spp. unravelled functional attributes. Genomics 112:1956–1969. https://doi.org/10.1016/j.ygeno.2019.11.008
Sood U, Hira P, Kumar R, Bajaj A, Rao DLN, Lal R, Shakarad M (2019) Comparative genomic analyses reveal core-genome-wide genes under positive selection and major regulatory hubs in outlier strains of Pseudomonas aeruginosa. Front Microbiol 10:53–76. https://doi.org/10.3389/fmicb.2019.00053
Kumar R, Verma H, Haider S, Bajaj A, Sood U, Ponnusamy K, Nagar S, Shakarad M, Negi RK, Singh Y, Khurana JP, Gilbert JA, Lal R (2017) Comparative genomic analysis reveals habitat-specific genes and regulatory hubs within the genus Novosphingobium. mSystems 2:e00020-17. https://doi.org/10.1128/mSystems.00020-17
Acknowledgements
RL acknowledges The National Academy of Sciences, India, for support under the NASI‐Senior Scientist Platinum Jubilee Fellowship Scheme. NS acknowledges Council of Scientific and Industrial Research, New Delhi (CSIR) for doctoral fellowship.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Singhvi, N., Gupta, V., Singh, P. et al. Prediction of Transcription Factors and Their Involvement in Regulating Rifamycin Production in Amycolatopsis mediterranei S699. Indian J Microbiol 60, 310–317 (2020). https://doi.org/10.1007/s12088-020-00868-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12088-020-00868-5