Abstract
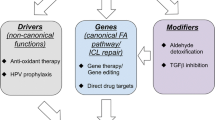
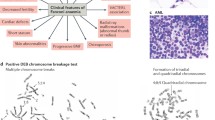
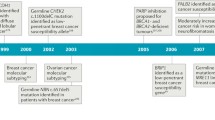
Breast cancer is one of the most frequent cancers in the world. The majority of cases are sporadic but around 15% show some type of familial aggregation and about 5% exhibit a clear hereditary pattern. Common and rare low-moderate-penetrance genes, and high-penetrance genes are thought to explain the genetic susceptibility to the disease. Only around 20% of the inherited risk to breast cancer is explained by germline mutations in the known high-penetrance susceptibility genes BRCA1 and BRCA2. Mutations in genes such as TP53 and PTEN have also been linked with high risk for breast cancer within specific cancer syndromes and rare germline variants in genes such as CHEK2 and ATM have been found to confer modest risk to breast cancer. However, we can say that less than 30% of familial risk of breast cancer is due to known genes. Identification in 2002 of the Fanconi anaemia (FA) gene FANCD1 as BRCA2 and recent studies indicating that heterozygous mutations in FANCN/PALB2 and FANCJ/ BRIP1 predispose to breast cancer have emphasised an important connection between the FA and BRCA pathway. Here we review the emerging DNA-damage response network consisting of FA and BRCA proteins, summarise what is currently known about the direct involvement of these molecules in breast cancer susceptibility and discuss the prospect offered by this pathway in order to identify more breast cancer related genes. We finally present the current stage of therapeutic options specifically targeting the FA/BRCA pathway and summarise the challenges this field encounters
Similar content being viewed by others
References
Lopez-Abente G, Pollan M, Aragones N et al (2004) [State of cancer in Spain: incidence]. An Sist Sanit Navar 27:165–173
Houlston RS, Peto J (2004) The search for low-penetrance cancer susceptibility alleles. Oncogene 23:6471–6476
Easton DF, Pooley KA, Dunning AM et al (2007) Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 447: 1087–1093
Stacey SN, Manolescu A, Sulem P et al (2007) Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet 39:865–869
Milne RL, Ribas G, Gonzalez-Neira A et al (2006) ERCC4 associated with breast cancer risk: a two-stage case-control study using high-throughput genotyping. Cancer Res 66:9420–9427
Thompson D, Antoniou AC, Jenkins M et al (2005) Two ATM variants and breast cancer risk. Hum Mutat 25:594–595
Meijers-Heijboer H, van den Ouweland A, Klijn J et al (2002) Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in non-carriers of BRCA1 or BRCA2 mutations. Nat Genet 31:55–59
Johnson N, Fletcher O, Palles C et al (2007) Counting potentially functional variants in BRCA1, BRCA2 and ATM predicts breast cancer susceptibility. Hum Mol Genet 16:1051–1057
Antoniou AC, Pharoah PD, McMullan G et al (2001) Evidence for further breast cancer susceptibility genes in addition to BRCA1 and BRCA2 in a population-based study. Genet Epidemiol 21:1–18
Diez O, Osorio A, Duran M et al (2003) Analysis of BRCA1 and BRCA2 genes in Spanish breast/ovarian cancer patients: a high proportion of mutations unique to Spain and evidence of founder effects. Hum Mutat 22:301–312
Ford D, Easton DF (1995) The genetics of breast and ovarian cancer. Br J Cancer 72:805–812
Chen S, Parmigiani G (2007) Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol 25:1329–1333
Antoniou A, Pharoah PD, Narod S et al (2003) Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet 72:1117–1130
García MJ, Fernández V, Osorio A, et al. Analysis of FANCB and FANCN/ PALB2 Fanconi Anemia genes in BRCA1/2-negative Spanish breast cancer families (submitted)
Venkitaraman AR (2002) Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108:171–182
Yoshida K, Miki Y (2004) Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription, and cell cycle in response to DNA damage. Cancer Sci 95:866–871
Kim H, Chen J, Yu X (2007) Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 316:1202–1205
Sobhian B, Shao G, Lilli DR et al (2007) RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316:1198–1202
Wang B, Matsuoka S, Ballif BA et al (2007) Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 316:1194–1198
Lu H, Guo X, Meng X et al (2005) The BRCA2-interacting protein BCCIP functions in RAD51 and BRCA2 focus formation and homologous recombinational repair. Mol Cell Biol 25:1949–1957
Joenje H, Patel KJ (2001) The emerging genetic and molecular basis of Fanconi anaemia. Nat Rev Genet 2:446–457
Hirano S, Yamamoto K, Ishiai M et al (2005) Functional relationships of FANCC to homologous recombination, translesion synthesis, and BLM. EMBO J 24:418–427
Akkari YM, Bateman RL, Reifsteck CA et al (2000) DNA replication is required to elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol Cell Biol 20:8283–8289
Wang W (2007) Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet 8:735–748
Callen E, Casado JA, Tischkowitz MD et al (2005) A common founder mutation in FANCA underlies the world’s highest prevalence of Fanconi anemia in Gypsy families from Spain. Blood 105:1946–1949
Verlander PC, Kaporis A, Liu Q et al (1995) Carrier frequency of the IVS4 + 4 A→T mutation of the Fanconi anemia gene FAC in the Ashkenazi Jewish population. Blood 86:4034–4038
Strathdee CA, Gavish H, Shannon WR, Buchwald M (1992) Cloning of cDNAs for Fanconi’s anaemia by functional complementation. Nature 358:434
Medhurst AL, Huber PA, Waisfisz Q et al (2001) Direct interactions of the five known Fanconi anaemia proteins suggest a common functional pathway. Hum Mol Genet 10:423–429
Timmers C, Taniguchi T, Hejna J et al (2001) Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol Cell 7:241–248
Sims AE, Spiteri E, Sims RJ 3rd et al (2007) FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol 14:564–567
Dorsman JC, Levitus M, Rockx D et al (2007) Identification of the Fanconi anemia complementation group I gene, FANCI. Cell Oncol 29:211–218
Howlett NG, Taniguchi T, Olson S et al (2002) Biallelic inactivation of BRCA2 in Fanconi anemia. Science 297:606–609
Alter BP, Rosenberg PS, Brody LC (2007) Clinical and molecular features associated with biallelic mutations in FANCD1/BRCA2. J Med Genet 44:1–9
Seal S, Barfoot R, Jayatilake H et al (2003) Evaluation of Fanconi Anemia genes in familial breast cancer predisposition. Cancer Res 63:8596–8599
Seal S, Thompson D, Renwick A et al (2006) Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet 38:1239–1241
Xia B, Sheng Q, Nakanishi K et al (2006) Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell 22:719–729
Xia B, Dorsman JC, Ameziane N et al (2007) Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet 39:159–161
Reid S, Schindler D, Hanenberg H et al (2007) Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet 39:162–164
Rahman N, Seal S, Thompson D et al (2007) PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet 39:165–167
Tischkowitz M, Xia B, Sabbaghian N et al (2007) Analysis of PALB2/FANCN-associated breast cancer families. Proc Natl Acad Sci U S A 104: 6788–6793
Milne RL, Osorio A, Ramón y Cajal T, et al. The average cumulative risks of breast and ovarian cancer for carriers of mutations in BRCA1 and BRCA2 attending genetic counselling units in Spain. Clinical Cancer Research (in press)
Erkko H, Xia B, Nikkila J et al (2007) A recurrent mutation in PALB2 in Finnish cancer families. Nature 446:316–319
Kim H, Huang J, Chen J (2007) CCDC98 is a BRCA1-BRCT domain-binding protein involved in the DNA damage response. Nat Struct Mol Biol 14:710–715
van der Heijden MS, Yeo CJ, Hruban RH, Kern SE (2003) Fanconi anemia gene mutations in young-onset pancreatic cancer. Cancer Res 63: 2585–2588
van der Heijden MS, Brody JR, Gallmeier E et al (2004) Functional defects in the Fanconi anemia pathway in pancreatic cancer cells. Am J Pathol 165: 651–657
Couch FJ, Johnson MR, Rabe K et al (2005) Germ line Fanconi anemia complementation group C mutations and pancreatic cancer. Cancer Res 65:383–386
Taniguchi T, Tischkowitz M, Ameziane N et al (2003) Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat Med 9:568–574
Narayan G, Arias-Pulido H, Nandula SV et al (2004) Promoter hypermethylation of FANCF: disruption of Fanconi Anemia-BRCA pathway in cervical cancer. Cancer Res 64:2994–2997
Marsit CJ, Liu M, Nelson HH et al (2004) Inactivation of the Fanconi anemia/BRCA pathway in lung and oral cancers: implications for treatment and survival. Oncogene 23:1000–1004
Jacinto FV, Esteller M (2007) Mutator pathways unleashed by epigenetic silencing in human cancer. Mutagenesis 22:247–253
Rio PG, Pernin D, Bay JO et al (1998) Loss of heterozygosity of BRCA1, BRCA2 and ATM genes in sporadic invasive ductal breast carcinoma. Int J Oncol 13:849–853
Katsama A, Sourvinos G, Zachos G, Spandidos DA (2000) Allelic loss at the BRCA1, BRCA2 and TP53 loci in human sporadic breast carcinoma. Cancer Lett 150:165–170
Gallmeier E, Kern SE (2007) Targeting Fanconi anemia/BRCA2 pathway defects in cancer: the significance of preclinical pharmacogenomic models. Clin Cancer Res 13:4–10
Kennedy RD, Chen CC, Stuckert P et al (2007) Fanconi anemia pathway-deficient tumor cells are hypersensitive to inhibition of ataxia telangiectasia mutated. J Clin Invest 117:1440–1449
Gallmeier E, Hucl T, Brody JR et al (2007) Highthroughput screening identifies novel agents eliciting hypersensitivity in Fanconi pathway-deficient cancer cells. Cancer Res 67:2169–2177
van der Heijden MS, Brody JR, Dezentje DA et al (2005) In vivo therapeutic responses contingent on Fanconi anemia/BRCA2 status of the tumor. Clin Cancer Res 11:7508–7515
Nguewa PA, Fuertes MA, Cepeda V et al (2006) Poly(ADP-ribose) polymerase-1 inhibitor 3-aminobenzamide enhances apoptosis induction by platinum complexes in cisplatin-resistant tumor cells. Med Chem 2:47–53
Bryant HE, Schultz N, Thomas HD et al (2005) Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434:913–917
Farmer H, McCabe N, Lord CJ et al (2005) Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434:917–921
Schreiber V, Ame JC, Dolle P et al (2002) Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J Biol Chem 277:23028–23036
Boulton S, Kyle S, Durkacz BW (1999) Interactive effects of inhibitors of poly(ADP-ribose) polymerase and DNA-dependent protein kinase on cellular responses to DNA damage. Carcinogenesis 20:199–203
Ratnam K, Low JA (2007) Current development of clinical inhibitors of poly(ADP-ribose) polymerase in oncology. Clin Cancer Res 13:1383–1388
Rubinstein WS (2007) Hereditary breast cancer: pathobiology, clinical translation, and potential for targeted cancer therapeutics. Fam Cancer. 2007 Jul 12; (Epub ahead of print)
Melchor L, Honrado E, Garcia MJ, Alvarez S, Palacios J, Osorio A, Nathanson KL, Benitez J (2007) Distinct genomic aberration patterns are found in familial breast cancer associated with different immunohistochemical subtypes. Oncogene. 2007 Dec 10; (Epub ahead of print)
Honrado E, Osorio A, Milne RL et al (2007) Immunohistochemical classification of non-BRCA1/2 tumors identifies different groups that demonstrate the heterogeneity of BRCAX families. Mod Pathol 20:1298–1306
Sorlie T, Perou CM, Tibshirani R et al (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 98:10869–10874
Sorlie T, Tibshirani R, Parker J et al (2003) Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A 100:8418–8423
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported by an unrestricted educational grant from Pfizer.
Rights and permissions
About this article
Cite this article
García, M.J., Benítez, J. The Fanconi anaemia/BRCA pathway and cancer susceptibility. Searching for new therapeutic targets. Clin Transl Oncol 10, 78–84 (2008). https://doi.org/10.1007/s12094-008-0160-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-008-0160-6