Abstract
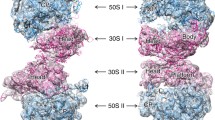
Staphylococcus aureus: hibernation-promoting factor (SaHPF) is a 22.2 kDa stationary-phase protein that binds to the ribosome and turns it to the inactive form favoring survival under stress. Sequence analysis has shown that this protein is combination of two homolog proteins obtained in Escherichia coli—ribosome hibernation promoting factor (HPF) (11,000 Da) and ribosome modulation factor RMF (6500 Da). Binding site of E. coli HPF on the ribosome have been shown by X-ray study of Thermus thermophilus ribosome complex. Hence, recent studies reported that the interface is markedly different between 100S from S. aureus and E. coli. Cryo-electron microscopy structure of 100S S. aureus ribosomes reveal that the SaHPF-NTD binds to the 30S subunit as observed for shorter variants of HPF in other species and the C-terminal domain (CTD) protrudes out of each ribosome in order to mediate dimerization. SaHPF-NTD binds to the small subunit similarly to its homologs EcHPF, EcYfiA, and a plastid-specific YfiA. Furthermore, upon binding to the small subunit, the SaHPF-NTD occludes several antibiotic binding sites at the A site (hygromycin B, tetracycline), P site (edeine) and E site (pactamycin, kasugamycin). In order to elucidate the structure, dynamics and function of SaHPF-NTD from S. aureus, here we report the backbone and side chain resonance assignments for SaHPF-NTD. Analysis of the backbone chemical shifts by TALOS+ suggests that SaHPF-NTD contains two α-helices and four β-strands (β1-α1-β2-β3-β4-α2 topology). Investigating the long-term survival of S. aureus and other bacteria under antibiotic pressure could lead to advances in antibiotherapy.


Similar content being viewed by others
Abbreviations
- IPTG:
-
Isopropyl-thio-β-d-galactoside
- S. aureus :
-
Staphylococcus aureus
- RMF:
-
Ribosome modulation factor
- SaHPF:
-
Staphylococcus aureus hibernation promoting factor
- SaHPF-NTD:
-
N-terminal domain of SaHPF
- SaHPF-CTD:
-
C-terminal domain of SaHPF
- Cryo-EM:
-
Cryo-electron microscopy
- PIC:
-
Protease inhibitor cocktail
- PMSF:
-
Phenylmethane sulfonyl fluoride
References
Akanuma G, Kazo Y, Tagami K, Hiraoka H, Yano K, Suzuki S, Hanai R, Nanamiya H, Kato-Yamada Y, Kawamura F (2016) Ribosome dimerization is essential for the efficient regrowth of Bacillus subtilis. Microbiology 162(3):448–458
Akiyama T, Williamson KS, Schaefer R, Pratt Sh, Chang CB, Franklin MJ (2017) Resuscitation of Pseudomonas aeruginosa from dormancy requires hibernation promoting factor (PA4463) for ribosome preservation. PNAS 114:3204–3209. doi:10.1073/pnas.1700695114
Ayupov RK, Akberova NI (2016a) Prediction of the three-dimensional structure of the protein SaHPF and analysis of its molecular dynamics. Int J Pharm Technol 8:14548–14557
Ayupov RKh, Akberova NI (2016b) Analysis of the structure of SaHPF Staphylococcus aureus protein. Uchenye Zapiski Kazanskogo Universiteta Seriya Estestvennye Nauki 158:327–337
Ayupov RK, Khusainov ISh, Validov SZ, Yusupova GZ, Yusupov MM (2016c) Isolation and purification of Staphylococcus aureus hibernation promoting factor inactivating of the ribosome. Int J Pharm Technol 8:14392–143986
Basu A, Yap MNF (2016) Ribosome hibernation factor promotes Staphylococcal survival and differentially represses translation. Nucleic Acids Res 44(10):4881–4893
Berjanskii MV, Wishart DS (2005) A simple method to predict protein flexibility using secondary chemical shifts. J Am Chem Soc 127:14970–14971. doi:10.1021/ja054842f
Bieri P, Leibundgut M, Saurer M, Boehringer D, Ban N (2017) The complete structure of the chloroplast 70S ribosome in complex with translation factor pY. EMBO J 36:475–486. doi:10.15252/embj.201695959
Borovinskaya MA, Shoji S, Fredrick K, Cate JH (2008) Structural basis for hygromycin B inhibition of protein biosynthesis. RNA 14:1590–1599. doi:10.1261/rna.1076908
Brodersen DE, Clemons WM Jr, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V (2000) The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 103:1143–1154
Daragan VA, Mayo KH (1997) Motional model analyses of protein and peptide dynamics using 13C and 15N NMR relaxation. Prog NMR Spectrosc 31:63–105
Jenner L et al (2013) Structural basis for potent inhibitory activity of the antibiotic tigecycline during protein synthesis. Proc Nat Acad Sci USA 110:3812–3816. doi:10.1073/pnas.1216691110
Kato T, Yoshida H, Miyata T, Maki Y, Wada A, Namba K (2010) Structure of the 100S ribosome in the hibernation stage revealed by electron cryomicroscopy. Structure 18(6):719–724
Khusainov I et al (2016) Structure of the 70S ribosome from human pathogen Staphylococcus aureus. Nucl Acids Res 44:10491–10504. doi:10.1093/nar/gkw1126
Khusainov I, Vicens Q, Ayupov R, Usachev K, Myasnikov A, Simonetti A, Validov Sh, Kieffer B, Yusupova G, Yusupov M, Hashem Ya (2017) Structures and dynamics of hibernating ribosomes from S. aureus mediated by intermolecular interactions of HPF. EMBO J 36(14):2073–2087. doi:10.15252/embj.201696105
McKay SL, Portnoy DA (2015) Ribosome hibernation facilitates tolerance of stationary-phase bacteria to aminoglycosides. Antimicrob Agents Chemother 59(11):6992–6999
Ortiz JO, Brandt F, Matias VRF, Sennels L, Rappsilber J, Scheres SHW, Eibauer M, Hartl FU, Baumeister W (2010) Structure of hibernating ribosomes studied by cryoelectron tomography in vitro and in situ. J Cell Biol 190(4):613–621
Pioletti M et al (2001) Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J 20:1829–1839. doi:10.1093/emboj/20.8.1829
Polikanov YS, Blaha GM, Steitz TA (2012) How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science 336:915–918. doi:10.1126/science.1218538
Russell JB, Cook GM (1995) Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol Rev 59:48–62
Schluenzen F et al (2006) The antibiotic kasugamycin mimics mRNA nucleotides to destabilize tRNA binding and inhibit canonical translation initiation. Nat Struct Mol Biol 13:871–878. doi:10.1038/nsmb1145
Sharma MR et al (2010) PSRP1 is not a ribosomal protein, but a ribosome-binding factor that is recycled by the ribosome-recycling factor (RRF) and elongation factor G (EF-G). J Biol Chem 285:4006–4014. doi:10.1074/jbc.M109.062299
Shen Y, Delaglio F, Cornilescu G, Bax A (2009) TALOS plus: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR 44:213–223. doi:10.1007/s10858-009-9333-z
Szaflarski W, Nierhaus KH (2007) Question 7: optimized energy consumption for protein synthesis. Origins of life and evolution of the biosphere. J Int Soc Study Orig Life 37:423–428. doi:10.1007/s11084-007-9091-4
Ueta M, Yoshida H, Wada C, Baba T, Mori H, Wada A (2005) Ribosome binding proteins YhbH and YfiA have opposite functions during 100S formation in the stationary phase of Escherichia coli. Genes Cells 10(12):1103–1112
Ueta M, Ohniwa RL, Yoshida H, Maki Y, Wada C, Wada A (2008) Role of HPF (hibernation promoting factor) in translational activity in Escherichia coli. J Biochem 143(3):425–433
Ueta M, Wada C, Wada A (2010) Formation of 100S ribosomes in Staphylococcus aureus by the hibernation promoting factor homolog SaHPF. Genes Cells 15:43–58. doi:10.1111/j.1365-2443.2009.01364.x
Vila-Sanjurjo A, Schuwirth BS, Hau CW, Cate JH (2004) Structural basis for the control of translation initiation during stress. Nat Struct Mol Biol 11:1054–1059. doi:10.1038/nsmb850
Vranken WF et al (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59:687–696. doi:10.1002/prot.20449
Yoshida H, Wada A (2014) The 100S ribosome: ribosomal hibernation induced by stress. Wiley Interdiscipl Rev RNA 5:723–732. doi:10.1002/wrna.1242
Acknowledgements
This work was supported by the Russian Science Foundation (Grant 16-14-10014).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Usachev, K.S., Ayupov, R.K., Validov, S.Z. et al. NMR assignments of the N-terminal domain of Staphylococcus aureus hibernation promoting factor (SaHPF). Biomol NMR Assign 12, 85–89 (2018). https://doi.org/10.1007/s12104-017-9783-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-017-9783-2