Abstract
Many soil Actinobacteria are potent producers of extracellular enzymes decomposing lignocellulose. Four strains of Actinobacteria with a high potential to hydrolyse cellulose and hemicellulose were identified among environmental isolates. The strains were grown on raw lignocellulosic substrates (olive pomace, oat flakes, sawdust, and wheat straw) under submerged fermentation in a laboratory scale. Modified Melin Norkrans Medium amended with raw lignocellulosic substrates as carbon sources (0.5%) was used to enhance lignocellulosic biomass decomposition. Three strains belonged to the genus Streptomyces and one strain to the genus Mycobacterium. Annotation of genomes showed high proportion of genes encoding for carbohydrate-active enzymes in Streptomyces sp. GESEQ-4 (537, i.e. 6% of 8404 genes), Streptomyces sp. GESEQ-13 (351 (5%) of 6705 genes), Streptomyces sp. GESEQ-35 (608 (6%) of 9788 genes), and Mycolicibacterium fortuitum subsp. fortuitum GESEQ-9 (222 (3%) of 6405 genes). These included plant cell wall-degrading enzymes belonging to the families GH1, GH2, GH3, GH5, GH6, GH9, GH10, GH12, GH16, GH26, GH30, GH39, GH48, GH51, and GH74, of which GH1, GH2, GH3, GH5, GH6, and GH16 were found in all four genomes. Assays for cellulose and hemicellulose degrading extracellular enzymes confirmed the ability of the isolates to decompose cellulose and hemicellulose. The highest endo-cleaving enzyme activities were produced by the strain Steptomyces sp. GESEQ-4 DSM 106287. Our results provide new perspectives into the enzymatic array by which the Actinobacteria break down complex lignocellulosic biomass. It is crucial to assess the genome to determine enzyme function as well as the enzyme families responsible for the degradation process in Actinobacteria. The potential degradation functions for these actinobacterial strains were validated by testing their cellulolytic and hemicellulolytic activities with various lignocellulosic substrates.
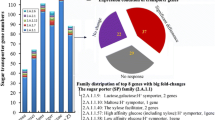
Graphic Abstract








Similar content being viewed by others
References
Ahmed, A.A.Q., Babalola, O.O., McKay, T.: Cellulase- and xylanase-producing bacterial isolates with the ability to saccharify wheat straw and their potential use in the production of pharmaceuticals and chemicals from lignocellulosic materials. Waste Biomass Valorization 9, 765–775 (2018). https://doi.org/10.1007/s12649-017-9849-5
Leo, V. V., Asem, D., Zothanpuia, Singh, B.P.: Actinobacteria: A highly potent source for holocellulose degrading enzymes. In: Singh, B.P., Gupta, V.K., Passari, A.K. (eds.) New and Future Developments in Microbial Biotechnology and Bioengineering: Actinobacteria: Diversity and Biotechnological Applications, pp. 191–205. Elsevier, Amsterdam (2018)
Větrovský, T., Steffen, K.T., Baldrian, P.: Potential of cometabolic transformation of polysaccharides and lignin in lignocellulose by soil Actinobacteria. PLoS ONE 9, e89108 (2014). https://doi.org/10.1371/journal.pone.0089108
López-Mondéjar, R., Algora, C., Baldrian, P.: Lignocellulolytic systems of soil bacteria: a vast and diverse toolbox for biotechnological conversion processes. Biotechnol. Adv. 37(6), 107374 (2019)
Kameshwar, S., Qin, W.: Recent developments in using advanced sequencing technologies for the genomic studies of lignin and cellulose degrading microorganisms. Int J Biol Sci. 12(2), 156 (2016). https://doi.org/10.7150/ijbs.13537
Anteneh, Y.S., Franco, C.M.M.: Whole cell actinobacteria as biocatalysts, www.frontiersin.org, (2019). https://doi.org/10.3389/fmicb.2019.00077
Blackman, L.M., Cullerne, D.P., Hardham, A.R.: Bioinformatic characterisation of genes encoding cell wall degrading enzymes in the Phytophthora parasitica genome. BMC Genomics (2014). https://doi.org/10.1186/1471-2164-15-785
Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P.M., Henrissat, B.: The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. (2014). https://doi.org/10.1093/nar/gkt1178
Anderson, I., Abt, B., Lykidis, A., Klenk, H.-P., Kyrpides, N., Ivanova, N.: Genomics of aerobic cellulose utilization systems in Actinobacteria. PLoS ONE 7, e39331 (2012). https://doi.org/10.1371/journal.pone.0039331
Cui, J., Mai, G., Wang, Z., Liu, Q., Zhou, Y., Ma, Y., Liu, C.: Metagenomic insights into a cellulose-rich niche reveal microbial cooperation in cellulose degradation. Front. Microbiol. (2019). https://doi.org/10.3389/fmicb.2019.00618
Küster, E., Williams, S.T.: Selection of media for isolation of streptomycetes. Nature 202, 928–929 (1964). https://doi.org/10.1038/202928a0
Hayakawa, M., Nonomura, H.: Humic acid-vitamin agar, a new medium for the selective isolation of soil actinomycetes. J. Ferment. Technol. 65, 501–509 (1987). https://doi.org/10.1016/0385-6380(87)90108-7
Houfani, A.A., Větrovský, T., Baldrian, P., Benallaoua, S.: Efficient screening of potential cellulases and hemicellulases produced by Bosea sp. FBZP-16 using the combination of enzyme assays and genome analysis. World J. Microbiol. Biotechnol. 33, 1–14 (2017). https://doi.org/10.1007/s11274-016-2198-x
Baldrian, P.: Microbial enzyme-catalyzed processes in soils and their analysis. Plant Soil Environ. 55, 370–378 (2009). https://doi.org/10.1007/s11104-008-9731-0
Lane, D.J.: 16S/23S rRNA sequencing. In: Stackebrandt, E.G., Goodfellow, M. (eds.) Nucleic Acid Techniques in Bacterial Systematics, pp. 115–175. Wiley, New York (1991)
Zerbino, D.R., Birney, E.: Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 (2008). https://doi.org/10.1101/gr.074492.107
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A.A., Dvorkin, M., Kulikov, A.S., Lesin, V.M., Nikolenko, S.I., Pham, S.O.N., Prjibelski, A.D., Pyshkin, A. V, Sirotkin, A. V, Vyahhi, N., Tesler, G., Alekseyev, M.A.X.A., Pevzner, P.A.: SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012). https://doi.org/10.1089/cmb.2012.0021
Meyer, F., Paarmann, D., D’Souza, M., Olson, R., Glass, E., Kubal, M., Paczian, T., Rodriguez, A., Stevens, R., Wilke, A., Wilkening, J., Edwards, R.: The metagenomics RAST server: a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 9, 1–8 (2008). https://doi.org/10.1002/9781118010518.ch37
Yin, Y., Mao, X., Yang, J., Chen, X., Mao, F., Xu, Y.: DbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 40, 445–451 (2012). https://doi.org/10.1093/nar/gks479
Seemann, T.: Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014). https://doi.org/10.1093/bioinformatics/btu153
Lee, M.D.: GToTree: a user-friendly workflow for phylogenomics. Bioinformatics 35, 4162–4164 (2019). https://doi.org/10.1093/bioinformatics/btz188
Hyatt, D., Chen, G.L., LoCascio, P.F., Land, M.L., Larimer, F.W., Hauser, L.J.: Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 11, 119 (2010). https://doi.org/10.1186/1471-2105-11-119
Eddy, S.R.: Accelerated profile HMM searches. PLoS Comput. Biol. 7, 1002195 (2011). https://doi.org/10.1371/journal.pcbi.1002195
Edgar, R.C.: MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). https://doi.org/10.1093/nar/gkh340
Capella-Gutierrez, S., Silla-Martinez, J.M., Gabaldon, T.: trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009). https://doi.org/10.1093/bioinformatics/btp348
Price, M.N., Dehal, P.S., Arkin, A.P.: FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 (2010). https://doi.org/10.1371/journal.pone.0009490
Tange, O.: GNU Parallel 2018. (2018). https://doi.org/10.5281/ZENODO.1146014
Letunic, I., Bork, P.: Interactive Tree of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39, W475–W478 (2011). https://doi.org/10.1093/nar/gkr201
Lacombe-Harvey, M.-È., Brzezinski, R., Beaulieu, C.: Chitinolytic functions in actinobacteria: ecology, enzymes, and evolution. Appl. Microbiol. Biotechnol. 102, 7219–7230 (2018). https://doi.org/10.1007/s00253-018-9149-4
Sharma, H.K., Xu, C., Qin, W.: Biological Pretreatment of Lignocellulosic Biomass for Biofuels and Bioproducts: An Overview. https://www.bp.com/ (2019)
Riyadi, F.A., Tahir, A.A., Yusof, N., Sabri, N.S.A., Noor, M.J.M.M., Akhir, F.N.M.D., Othman, N., Zakaria, Z., Hara, H.: Enzymatic and genetic characterization of lignin depolymerization by Streptomyces sp. S6 isolated from a tropical environment. Sci. Rep. 10, 1–9 (2020). https://doi.org/10.1038/s41598-020-64817-4
Shrestha, S., Kognou, A.L.M., Zhang, J., Qin, W.: Different facets of lignocellulosic biomass including pectin and its perspectives. Waste Biomass Valorization 12, 4805–4823 (2020). https://doi.org/10.1007/s12649-020-01305-w
Aakko, J., Pietilä, S., Toivonen, R., Rokka, A., Mokkala, K., Laitinen, K., Elo, L., Hänninen, A.: A carbohydrate-active enzyme (CAZy) profile links successful metabolic specialization of Prevotella to its abundance in gut microbiota. Sci. Rep. 10, 12411 (2020). https://doi.org/10.1038/s41598-020-69241-2
Boutard, M., Cerisy, T., Nogue, P.-Y., Alberti, A., Weissenbach, J., Salanoubat, M., Tolonen, A.C.: Functional diversity of carbohydrate-active enzymes enabling a bacterium to ferment plant biomass. PLoS Genet. 10, e1004773 (2014). https://doi.org/10.1371/journal.pgen.1004773
Lladó Fernández, S., Větrovský, T., Baldrian, P.: The concept of operational taxonomic units revisited: genomes of bacteria that are regarded as closely related are often highly dissimilar. Folia Microbiol. (Praha) 64, 19–23 (2019). https://doi.org/10.1007/s12223-018-0627-y
Berlemont, R., Martiny, A.C.: Genomic potential for polysaccharide deconstruction in bacteria. Appl. Environ. Microbiol. 81, 1513–1519 (2015). https://doi.org/10.1128/AEM.03718-14
Berlemont, R., Martiny, A.C.: Phylogenetic distribution of potential cellulases in bacteria. Appl. Environ. Microbiol. 79, 1545–1554 (2013). https://doi.org/10.1128/AEM.03305-12
Ventura, M., Canchaya, C., Tauch, A., Chandra, G., Fitzgerald, G.F., Chater, K.F., van Sinderen, D.: Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71, 495–548 (2007). https://doi.org/10.1128/mmbr.00005-07
Malik, A., Kim, Y.R., Jang, I.H., Hwang, S., Oh, D.C., Kim, S.B.: Genome-based analysis for the bioactive potential of Streptomyces yeochonensis CN732, an acidophilic filamentous soil actinobacterium. BMC Genomics 21, 118 (2020). https://doi.org/10.1186/s12864-020-6468-5
Acknowledgements
The authors acknowledge the financial support by the Algerian Ministry of Higher Education and Scientific Research and the General Direction for Scientific Research and Technological Development (Algeria). The authors acknowledge Lamia Medouni–Haroune (Laboratoire de Microbiologie Appliquée, Université de Bejaia) and Karel Švec (Laboratory of Fungal Genetics and Metabolism, Institute of Microbiology of the Czech Academy of Sciences) for providing olive pomace and oat flakes, respectively.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Houfani, A.A., Tláskal, V., Baldrian, P. et al. Actinobacterial Strains as Genomic Candidates for Characterization of Genes Encoding Enzymes in Bioconversion of Lignocellulose. Waste Biomass Valor 13, 1523–1534 (2022). https://doi.org/10.1007/s12649-021-01595-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12649-021-01595-8