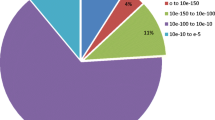
Abstract
Saccharina is one of the most important cold-water living marine brown algal genera. In this study we analyzed the transcriptome of S. japonica, which belongs to the 1 000 Plants (OneKP) Project, by using a next-generation high-throughput DNA sequencing technique. About 5.16 GB of raw data were generated, and 65 536 scaffolds with an average length of 454 bp were assembled with SOAP de novo assembly method. In total, 19 040 unigenes were identified by BLAST; 25 734 scaffolds were clustered into 37 Gene ontology functional groups; 6 760 scaffolds were classified into 25 COG categories, as well as 2 665 scaffolds that were assigned to 306 KEGG pathways. Majority of the unigenes exhibited more similarities to algae including brown algae and diatom than other cyanobacteria, marine diatom, and plant. Saccharina japonica has the outstanding capability to accumulate halogen such as Br and I via halogenation processes from seawater. We acquired 42 different vanadium-dependent haloperoxidases (vHPO) in S. japonica transcriptome data, including 5 segments of vanadium-dependent iodoperoxidase (vIPO) and 37 segments of vanadium-dependent bromoperoxidase (vBPO). Complicated analyses of identified fulllength S. japonica vBPO1 and S. japonica vBPO2 revealed the importance of vBPO among species of brown algae and the strong relationship between marine algal vBPOs and vIPOs. This study will enhance our understanding of the biological characteristics and economic values of S. japonica species.
Similar content being viewed by others
References
Almeida M, Humanes M, Melo R, et al. 1998. Sacchoriza polyschides (phaeophyceae; phyllariaceae): a new source for vanadium-dependent haloperoxidases. Phytochemistry, 48(2): 229–239
Almeida M G, Humanes M, Melo R, et al. 2000. Purification and characterization of vanadium haloperoxidases from the brown alga Pelvetia canaliculata. Phytochemistry, 54(1): 5–11
Altermann E, Klaenhammer T R. 2005. PathwayVoyager: pathway mapping using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. BMC Genomics, 6: 60
Baden D G, Corbett M D. 1980. Bromoperoxidases from Penicillus capitatus, Penicillus lamourouxii and Rhipocephalus phoenix. Biochem. J 187(1): 205–211
Bernroitner M, Zamocky M, Furtmüller P G, et al. 2009. Occurrence, phylogeny, structure, and function of catalases and peroxidases in cyanobacteria. J Exp Bot, 60(2): 423–440
Blobel G, Dobberstein B. 1975. Transfer of proteins across membranes: I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Bio, 67(3): 835–851
Bold H C, Wynne M J. 1985. Introduction to the Algae: Structure and Reproduction. Engelwood Cliffs: Prentice Hall Inc
Butler A, Carter-Franklin J N. 2004. The role of vanadium bromoperoxidase in the biosynthesis of halogenated marine natural products. Nat Prod Rep, 21(1): 180–188
Butler A, Walker J V. 1993. Marine Haloperoxidases. Chem Rev, 93: 1937–1994
Colin C, Leblanc C, Wagner E, et al. 2003. The brown algal kelp Laminaria digitata features distinct bromoperoxidase and iodoperoxidase activities. J Biol Chem, 278(26): 23545–23552
Conesa A, Gotz S. 2008. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics, 2008: 619832
Dembitsky V M. 2003. Oxidation, epoxidation and sulfoxidation reactions catalysed by haloperoxidases. Tetrahedron, 59(26): 4701–4720
Deng Yunyan, Yao Jianting, Wang Xiuliang, et al. 2012. Transcriptome sequencing and comparative analysis of Saccharina japonica (Laminariales, Phaeophyceae) under blue light induction. PLoS One, 7(6): e39704
Ghangal R, Raghuvanshi S, Sharma P C. 2009. Isolation of good quality RNA from a medicinal plant seabuckthorn rich in secondary metabolites. Plant Physiol Biochem, 47(11–12): 1113–1115
Graham L E, Wilcox L W. 2000. Algae. Prentice Hall, NJ: Upper Saddle River
Gray M W. 2012. Mitochondrial evolution. Cold Spring Harb Perspect Biol, 4(9): a011403
Hemrika W, Renirie R, Dekker H L, et al. 1997. From phosphatases to vanadium peroxidases: a similar architecture of the active site. Proc Natl Acad Sci USA, 94(6): 2145–2149
Kanehisa M, Goto S, Kawashima S, et al. 2004. The KEGG resource for deciphering the genome. Nucleic Acids Res, 32(Database issue): D277–280
Kawai H, Sasaki H. 2000. Molecular phylogeny of the brown algal genera Akkesiphycus and AkkesiphycusHalosiphon (Laminariales), resulting in the circumscription of the new families Akkesiphycaceae and Halosiphonaceae. Phycologia, 39(5): 416–428
Li Ruiqiang, Li Yingrui, Kristiansen K, et al. 2008. SOAP: short oligonucleotide alignment program. Bioinformatics, 24(5): 713–714
Li Tianyong, Ren Lei. 2012. A suitable method for extracting total RNA from red algae. Transactions of Oceanology and Limnology (in Chinese), 4: 64–71
Li Chaozheng, Weng Shaoping, Chen Yonggui, et al. 2012. Analysis of Litopenaeus vannamei transcriptome using the next-generation DNA sequencing technique. PLoS One, 7(10): e47442
Littlechild J, Garcia-Rodriguez E, Dalby A, et al. 2002. Structural and functional comparisons between vanadium haloperoxidase and acid phosphatase enzymes. J Mol Recognit, 15(5): 291–296
Moriya Y, Itoh M, Okuda S, et al. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Research, 35(Web Server Issue): W182–185
Neuwald A F. 1997. An unexpected structural relationship between integral membrane phosphatases and soluble haloperoxidases. Protein Sci, 6(8): 1764–1767
Ogata H, Goto S, Sato K, et al. 1999. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res, 27(1): 29–34
Page R D. 1996. TREEVIEW: An application to display phylogenetic trees on personal computers. Comput Appl Biosci, 12(4): 357–358
Ronquist F, Huelsenbeck J P. 2003. Mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19(12): 1572–1574
Saenko G N, Kravtsova Y Y, Ivaneneko V V, et al. 1978. Concentration of iodine and bromine by plants in the Seas of Japan and Okhotsk. Marine Biology, 47: 243–250
Shimonishi M, Kuwamoto S, Inoue H, et al. 1998. Cloning and expression of the gene for a vanadium-dependent bromoperoxidase from a marine macro-alga, Corallina pilulifera. FEBS Letters, 428(1–2): 105–110
Sonnhammer E L, von Heijne G, Krogh A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol, 6: 175–182
Tatusov R L, Fedorova N D, Jackson J D, et al. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics, 4: 41
Petersen T N, Brunak S, von Heijne G, et al. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods, 8(10): 785–786
Thompson J D, Gibson T J, Plewniak F, et al. 1997. The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res, 25(24): 4876–4882
Tseng C K. 2001. Algal biotechnology industries and research activities in China. Journal of Applied Phycology, 13: 375–380
Van den Hoek C, Mann D G, Jahns H M. 1995. Algae: An Introduction to Phycology. Cambridge: Cambridge University Press
Vilter H. 1984. Peroxidases from Phaeophyceae: a vanadium(V)-dependent peroxidase from Ascophyllum nodosum. Phytochemistry, 23(7): 1387–1390
Vilter H. 1995. Vanadium-dependent haloperoxidases. Met Ions Biol Syst, 31: 325–362
Wang Wenjun, Wang Feijiu, Sun Xiutao, et al. 2013. Comparison of transcriptome under red and blue light culture of Saccharina japonica (Phaeophyceae). Planta J, 237(4): 1123–1133
Wever R, Tromp M G M, Krenn B E, et al. 1991. Brominating activity of the seaweed Ascophyllum nodosum: impact on the biosphere. Environ Sci Technol, 25(3): 446–449
Weyand M, Hecht H, Kiess M, et al. 1999. X-ray structure determination of a vanadium-dependent haloperoxidase from Ascophyllum nodosum at 2.0 A resolution. J Mol Biol, 293(3): 595–611
Weyand M, Schlichting I, Marabotti A, et al. 2002. Crystal structures of a new class of allosteric effectors complexed to tryptophan synthase. J Biol Chem, 277(12): 10647–10652
Winter J M, Moore B S. 2009. Exploring the chemistry and biology of vanadium-dependent haloperoxidases. J Biol Chem, 284(28): 18577–18581
Wixon J, Kell D. 2000. The Kyoto encyclopedia of genes and genomes-KEGG. Yeast, 17: 48–55
Xu Jia, Aileni M, Abbagani S, et al. 2010. A reliable and efficient method for total RNA isolation from various members of Spurge Family (Euphorbiaceae). Phytochem Anal, 21(5): 395–398
Yao Jianting, Fu Wandong, Wang Xiuliang, et al. 2009. Improved RNA isolation from Laminaria japonica Aresch (Laminariaceae, Phaeophyta). J Appl Phycol, 21: 233–238
Ye Jia, Fang Lin, Zheng Hongkun, et al. 2006. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res, 34(Web Server issue): W293–297
Zdobnov E M, Apweiler R. 2001. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics, 17(9): 847–848
Author information
Authors and Affiliations
Corresponding authors
Additional information
Foundation item: The National Natural Science Foundation of China under contract Nos 41206116, 31140070 and 31271397; Technology Project of Ocean and Fisheries of Guangdong Province under contract No. A201201E03; the Fundamental Research Funds for the Central Universities under contract No. 201262003; the algal transcriptome sequencing was supported by OneKP Project (www.onekp.com).
Rights and permissions
About this article
Cite this article
Liang, X., Wang, X., Chi, S. et al. Analysis of Saccharina japonica transcriptome using the high-throughput DNA sequencing technique and its vanadium-dependent haloperoxidase gene. Acta Oceanol. Sin. 33, 27–36 (2014). https://doi.org/10.1007/s13131-014-0438-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13131-014-0438-1