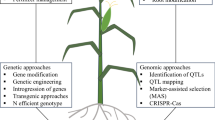
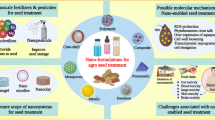
Abstract
Seeds serve as biochemical factories of nutrition, processing, bio-energy and storage related important bio-molecules and act as a delivery system to transmit the genetic information to the next generation. The research pertaining towards delineating the complex system of regulation of genes and pathways related to seed biology and nutrient partitioning is still under infancy. To understand these, it is important to know the genes and pathway(s) involved in the homeostasis of bio-molecules. In recent past with the advent and advancement of modern tools of genomics and genetic engineering, multi-layered ‘omics’ approaches and high-throughput platforms are being used to discern the genes and proteins involved in various metabolic, and signaling pathways and their regulations for understanding the molecular genetics of biosynthesis and homeostasis of bio-molecules. This can be possible by exploring systems biology approaches via the integration of omics data for understanding the intricacy of seed development and nutrient partitioning. These information can be exploited for the improvement of biologically important chemicals for large-scale production of nutrients and nutraceuticals through pathway engineering and biotechnology. This review article thus describes different omics tools and other branches that are merged to build the most attractive area of research towards establishing the seeds as biochemical factories for human health and nutrition.



Similar content being viewed by others
References
Agarwal P, Kapoor S, Tyagi AK (2011) Transcription factors regulating the progression of monocot and dicot seed development. Bioessays 33(3):189–202
Anderson A, Hudson M, Chen W, Zhu T (2003) Identification of nutrient partitioning genes participating in rice grain filling by singular value decomposition (SVD) of genome expression data. BMC Genom 4(1):26
Balestrazzi A, Confalonieri M, Macovei A, Donà M, Carbonera D (2011) Genotoxic stress and DNA repair in plants: emerging functions and tools for improving crop productivity. Plant Cell Rep 30(3):287–295
Bevan MW, Uauy C (2013) Genomics reveals new landscapes for crop improvement. Genome Biol 14(6):206
Bewley JD, Bradford K, Hilhorst H (2012) Seeds: physiology of development, germination and dormancy. Springer, Berlin
Brügger B (2014) Lipidomics: analysis of the lipid composition of cells and subcellular organelles by electrospray ionization mass spectrometry. Annu Rev Biochem 83:79–98
Butcher EC, Berg EL, Kunkel EJ (2004) Systems biology in drug discovery. Nat Biotechnol 22(10):1253
Cai J, Okamoto M, Atieno J, Sutton T, Li Y, Miklavcic SJ (2016) Quantifying the onset and progression of plant senescence by color image analysis for high throughput applications. PLoS One 11(6):e0157102
Collakova E, Aghamirzaie D, Fang Y, Klumas C, Tabataba F, Kakumanu A, Myers E, Heath LS, Grene R (2013) Metabolic and transcriptional reprogramming in developing soybean (Glycine max) embryos. Metabolites 3(2):347–372
Dam S, Thaysen-Andersen M, Stenkjær E, Lorentzen A, Roepstorff P, Packer NH, Stougaard J (2013) Combined N-glycome and N-glycoproteome analysis of the Lotus japonicus seed globulin fraction shows conservation of protein structure and glycosylation in legumes. J Proteome Res 12(7):3383–3392
Dean G, Cao Y, Xiang D, Provart NJ, Ramsay L, Ahad A, White R, Selvaraj G, Datla R, Haughn G (2011) Analysis of gene expression patterns during seed coat development in Arabidopsis. Mol Plant 4(6):1074–1091
Dekkers BJ, He H, Hanson J, Willems LA, Jamar DC, Cueff G, Rajjou L, Hilhorst HW, Bentsink L (2016) The Arabidopsis DELAY OF GERMINATION 1 gene affects ABSCISIC ACID INSENSITIVE 5 (ABI 5) expression and genetically interacts with ABI 3 during Arabidopsis seed development. Plant J 85(4):451–465
Deng ZY, Gong CY, Wang T (2013) Use of proteomics to understand seed development in rice. Proteomics 13(12–13):1784–1800
Diaz-Vivancos P, Faize M, Barba-Espin G, Faize L, Petri C, Hernández JA, Burgos L (2013) Ectopic expression of cytosolic superoxide dismutase and ascorbate peroxidase leads to salt stress tolerance in transgenic plums. Plant Biotechnol J 11(8):976–985
Diggle PK, Abrahamson NJ, Baker RL, Barnes MG, Koontz TL, Lay CR, Medeiros JS, Murgel JL, Shaner MG, Simpson HL, Wu CC (2010) Dynamics of maternal and paternal effects on embryo and seed development in wild radish (Raphanus sativus). Ann Bot 106(2):309–319
Dumas C, Rogowsky P (2008) Fertilization and early seed formation. Comptes Rendus Biol 331(10):715–725
Edlich-Muth C, Muraya MM, Altmann T, Selbig J (2016) Phenomic prediction of maize hybrids. Biosystems 146:102–109
Eldakak M, Milad SI, Nawar AI, Rohila JS (2013) Proteomics: a biotechnology tool for crop improvement. Front Plant Sci 4:35
El-Maarouf-Bouteau H, Meimoun P, Job C, Job D, Bailly C (2013) Role of protein and mRNA oxidation in seed dormancy and germination. Front Plant Sci 4:77
Engel R (2009) Development of analytical method for determination of water soluble vitamins in functional food product (Doctoral dissertation. Thesis of Ph. D. Dissertation Corvinus University of Budapest, Faculty of Food Science, Department of Applied Chemistry)
Fang W, Wang Z, Cui R, Li J, Li Y (2012) Maternal control of seed size by EOD3/CYP78A6 in Arabidopsis thaliana. Plant J 70(6):929–939
Figueiredo DD, Köhler C (2014) Signaling events regulating seed coat development. Biochem Soc Trans 42:358–363
Fukushima A, Kusano M (2013) Recent progress in the development of metabolome databases for plant systems biology. Front Plant Sci 4:73
Fukushima A, Kanaya S, Nishida K (2014) Integrated network analysis and effective tools in plant systems biology. Front Plant Sci 5:598
Furutani I, Sukegawa S, Kyozuka J (2006) Genome-wide analysis of spatial and temporal gene expression in rice panicle development. Plant J 46(3):503–511
Gajardo HA, Wittkop B, Soto-Cerda B, Higgins EE, Parkin IA, Snowdon RJ, Federico ML, Iniguez-Luy FL (2015) Association mapping of seed quality traits in Brassica napus L. using GWAS and candidate QTL approaches. Mol Breed 35(6):143
Gallardo K, Firnhaber C, Zuber H, Héricher D, Belghazi M, Henry C, Küster H, Thompson R (2007) A combined proteome and transcriptome analysis of developing medicago truncatula seeds evidence for metabolic specialization of maternal and filial tissues. Mol Cell Proteom 6(12):2165–2179
Gaur VS, Singh US, Kumar A (2011) Transcriptional profiling and in silico analysis of Dof transcription factor gene family for understanding their regulation during seed development of rice Oryza sativa L. Mol Biol Rep 38(4):2827–2848
Gehring JL, Delph LF (2006) Effects of reduced source-sink ratio on the cost of reproduction in females of Silene latifolia. Int J Plant Sci 167(4):843–851
Grant D, Nelson RT, Cannon SB, Shoemaker RC (2009) SoyBase, the USDA-ARS soybean genetics and genomics database. Nucl Acids Res 38(suppl_1):D843–D846
Grebe M, Gadea J, Steinmann T, Kientz M, Rahfeld JU, Salchert K, Koncz C, Jürgensa G (2000) A conserved domain of the Arabidopsis GNOM protein mediates subunit interaction and cyclophilin 5 binding. Plant Cell 12(3):343–356
Gu XY, Turnipseed EB, Foley ME (2008) The qSD12 locus controls offspring tissue-imposed seed dormancy in rice. Genetics 179(4):2263–2273
Gupta MK, Misra K (2016) A holistic approach for integration of biological systems and usage in drug discovery. Netw Model Anal Health Inform Bioinform 5(1):4
Gupta N, Gupta AK, Singh NK, Kumar A (2011) Differential expression of PBF Dof transcription factor in different tissues of three finger millet genotypes differing in seed protein content and color. Plant Mol Biol Rep 29(1):69–76
Gupta M, Bhaskar PB, Sriram S, Wang PH (2017) Integration of omics approaches to understand oil/protein content during seed development in oilseed crops. Plant Cell Rep 36(5):637–652
Gupta S, Pathak RK, Gupta SM, Gaur VS, Singh NK, Kumar A (2018) Identification and molecular characterization of Dof transcription factor gene family preferentially expressed in developing spikes of Eleusine coracana L.. 3 Biotech 8(2):82
Gurwitz D (2014) From transcriptomics to biological networks. Drug Dev Res 75(5):267–270
Gustin JL, Settles AM (2015) Seed phenomics. In: Phenomics. Springer, Berlin, pp 67–82
Hardtke CS, Berleth T (1998) The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J 17(5):1405–1411
He D, Yang P (2013) Proteomics of rice seed germination. Front Plant Sci 4:246
He H, Willems LA, Batushansky A, Fait A, Hanson J, Nijveen H, Hilhorst HW, Bentsink L (2016) Effects of parental temperature and nitrate on seed performance are reflected by partly overlapping genetic and metabolic pathways. Plant Cell Physiol 57(3):473–487
Hennig L, Derkacheva M (2009) Diversity of polycomb group complexes in plants: same rules, different players? Trends Genet 25(9):414–423
Hong M, Hu K, Tian T, Li X, Chen L, Zhang Y, Yi B, Wen J, Ma C, Shen J, Fu T (2017) Transcriptomic analysis of seed coats in yellow-seeded Brassica napus reveals novel genes that influence proanthocyanidin biosynthesis. Front Plant Sci 8:1674
Hori K, Sato K, Takeda K (2007) Detection of seed dormancy QTL in multiple mapping populations derived from crosses involving novel barley germplasm. Theor Appl Genet 115(6):869–876
Horn PJ, Chapman KD (2014) Lipidomics in situ: insights into plant lipid metabolism from high resolution spatial maps of metabolites. Progress Lip Res 54:32–52
Hu D, Tateno H, Hirabayashi J (2015) Lectin engineering, a molecular evolutionary approach to expanding the lectin utilities. Molecules 20(5):7637–7656
Hunt L, Bailey KJ, Gray JE (2010) The signalling peptide EPFL9 is a positive regulator of stomatal development. New Phytol 186(3):609–614
Huo H, Wei S, Bradford KJ (2016) DELAY OF GERMINATION1 (DOG1) regulates both seed dormancy and flowering time through microRNA pathways. Proc Natl Acad Sci 113(15):E2199–E2206
Hur M, Campbell AA, Almeida-de-Macedo M, Li L, Ransom N, Jose A, Crispin M, Nikolau BJ, Wurtele ES (2013) A global approach to analysis and interpretation of metabolic data for plant natural product discovery. Nat Prod Rep 30(4):565–583
Hwang EY, Song Q, Jia G, Specht JE, Hyten DL, Costa J, Cregan PB (2014) A genome-wide association study of seed protein and oil content in soybean. BMC Genom 15(1):1
Imtiaz M, Ogbonnaya FC, Oman J, van Ginkel M (2008) Characterization of quantitative trait loci controlling genetic variation for preharvest sprouting in synthetic backcross-derived wheat lines. Genetics 178(3):1725–1736
Ishibashi Y, Koda Y, Zheng SH, Yuasa T, Iwaya-Inoue M (2012) Regulation of soybean seed germination through ethylene production in response to reactive oxygen species. Ann Bot 111(1):95–102
Jiang H, Köhler C (2012) Evolution, function, and regulation of genomic imprinting in plant seed development. J Exp Bot 63(13):4713–4722
Jing L, Dombinov V, Shen S, Wu Y, Yang L, Wang Y, Frei M (2016) Physiological and genotype-specific factors associated with grain quality changes in rice exposed to high ozone. Environ Pollut 210:397–408
Jofuku KD, Den Boer BG, Van Montagu M, Okamuro JK (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6(9):1211–1225
Kazmi RH, Willems LA, Joosen RV, Khan N, Ligterink W, Hilhorst HW (2013) Metabolomic analysis of tomato seed germination. Metabolomics 13(12):145
Kiegle EA, Garden A, Lacchini E, Kater MM (2018) A genomic view of alternative splicing of long non-coding RNAs during rice seed development reveals extensive splicing and lncRNA gene families. Front Plant Sci 9:115
Kigel J (ed) (1995) Seed development and germination, vol 41. CRC Press, Boca Raton, pp 22–29 (ISBN 9780824792299)
Koller A, Washburn MP, Lange BM, Andon NL, Deciu C, Haynes PA, Hays L, Schieltz D, Ulaszek R, Wei J, Wolters D (2002) Proteomic survey of metabolic pathways in rice. Proc Natl Acad Sci 99(18):11969–11974
Körber N, Bus A, Li J, Parkin IA, Wittkop B, Snowdon RJ, Stich B (2016) Agronomic and seed quality traits dissected by genome-wide association mapping in Brassica napus. Front Plant Sci 7:386
Kranner I, Kastberger G, Hartbauer M, Pritchard HW (2010) Noninvasive diagnosis of seed viability using infrared thermography. Proc Natl Acad Sci 107(8):3912–3917
Kumar R, Taware R, Gaur VS, Guru SK, Kumar A (2009) Influence of nitrogen on the expression of TaDof1 transcription factor in wheat and its relationship with photo synthetic and ammonium assimilating efficiency. Mol Biol Rep 36(8):2209
Kumar A, Gaur VS, Goel A, Gupta AK (2015a) De novo assembly and characterization of developing spikes transcriptome of finger millet (Eleusine coracana): a minor crop having nutraceutical properties. Plant Mol Biol Rep 33(4):905–922
Kumar A, Pathak RK, Gupta SM, Gaur VS, Pandey D (2015b) Systems biology for smart crops and agricultural innovation: filling the gaps between genotype and phenotype for complex traits linked with robust agricultural productivity and sustainability. Omics J Integr Biol 19(10):581–601
Lariguet P, Ranocha P, De Meyer M, Barbier O, Penel C, Dunand C (2013) Identification of a hydrogen peroxide signalling pathway in the control of light-dependent germination in Arabidopsis. Planta 238(2):381–395
Le BH, Cheng C, Bui AQ, Wagmaister JA, Henry KF, Pelletier J, Kwong L, Belmonte M, Kirkbride R, Horvath S, Drews GN (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci 107(18):8063–8070
Li F, Wu X, Tsang E, Cutler AJ (2005) Transcriptional profiling of imbibed Brassica napus seed. Genomics 86(6):718–730
Li Y, Zheng L, Corke F, Smith C, Bevan MW (2008) Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev 22(10):1331–1336
Li J, Nie X, Tan JL, Berger F (2013) Integration of epigenetic and genetic controls of seed size by cytokinin in Arabidopsis. Proc Natl Acad Sci 110(38):15479–15484
Li L, Hur M, Lee JY, Zhou W, Song Z, Ransom N, Demirkale CY, Nettleton D, Westgate M, Arendsee Z, Iyer V (2015) A systems biology approach toward understanding seed composition in soybean. InBMC Genom 16(3):S9 (BioMed Central)
Locascio A, Roig-Villanova I, Bernardi J, Varotto S (2014) Current perspectives on the hormonal control of seed development in Arabidopsis and maize: a focus on auxin. Front Plant Sci 5:412
Lohe AR, Chaudhury A (2002) Genetic and epigenetic processes in seed development. Curr Opin Plant Biol 5(1):19–25
Long J, Barton MK (2000) Initiation of axillary and floral meristems in Arabidopsis. Dev Biol 218(2):341–353
Lu L, Tian S, Liao H, Zhang J, Yang X, Labavitch JM, Chen W (2013) Analysis of metal element distributions in rice (Oryza sativa L.) seeds and relocation during germination based on X-ray fluorescence imaging of Zn, Fe, K, Ca, and Mn. PLoS One 8(2):e57360
Macovei A, Pagano A, Leonetti P, Carbonera D, Balestrazzi A, Araújo SS (2017) Systems biology and genome-wide approaches to unveil the molecular players involved in the pre-germinative metabolism: implications on seed technology traits. Plant Cell Rep 36(5):669–688
Miernyk JA, Hajduch M (2011) Seed proteomics. J Proteom 74(4):389–400
Nadeau J, Lee E, Williams A, Oneill S (1995) Isolation of the arabidopsis homologs of 2 genes expressed in phalaenopsis ovules—a homeobox gene and a gene of unknown function. Plant Physiol 108:117–117
Navarro PJ, Pérez F, Weiss J, Egea-Cortines M (2016) Machine learning and computer vision system for phenotype data acquisition and analysis in plants. Sensors 16(5):641
Nodine MD, Bartel DP (2010) MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev 24(23):2678–2692
Nonogaki H (2017) Seed biology updates—highlights and new discoveries in seed dormancy and germination research. Front Plant Sci 8:524
Ogbonnaya FC, Imtiaz M, Ye G, Hearnden PR, Hernandez E, Eastwood RF, Van Ginkel M, Shorter SC, Winchester JM (2008) Genetic and QTL analyses of seed dormancy and preharvest sprouting resistance in the wheat germplasm CN10955. Theor Appl Genet 116(7):891–902
Ohnishi T, Sekine D, Kinoshita T (2014) Genomic imprinting in plants: what makes the functions of paternal and maternal genes different in endosperm formation? In: Advances in genetics, vol 86. Academic Press, London, pp 1–25
Palovaara J, Saiga S, Weijers D (2013) Transcriptomics approaches in the early Arabidopsis embryo. Trends Plant Sci 18(9):514–521
Pathak RK, Taj G, Pandey D, Arora S, Kumar A (2013) Modeling of the MAPK machinery activation in response to various abiotic and biotic stresses in plants by a system biology approach. Bioinformation 9(9):443
Pathak RK, Baunthiyal M, Pandey N, Pandey D, Kumar A (2017) Modeling of the jasmonate signaling pathway in Arabidopsis thaliana with respect to pathophysiology of Alternaria blight in Brassica. Sci Rep 7(1):16790
Pluskota WE, Martínez-Andújar C, Martin RC, Nonogaki H (2011) Non coding RNAs in plants. In: Erdmann VA, Barciszewski J, Pluskota WE, Martínez-Andújar C, Martin RC, Nonogaki H (eds) MicroRNA function in seed biology. Springer, Berlin
Prasad K, Zhang X, Tobón E, Ambrose BA (2010) The Arabidopsis B-sister MADS-box protein, GORDITA, represses fruit growth and contributes to integument development. Plant J 62(2):203–214
Raghavan V (2000) Developmental biology of flowering plants. Springer, New York
Rahaman M, Chen D, Gillani Z, Klukas C, Chen M (2015) Advanced phenotyping and phenotype data analysis for the study of plant growth and development. Front Plant Sci 10:619
Rajendran SR, Yau YY, Pandey D, Kumar A (2015) CRISPR-Cas9 based genome engineering: opportunities in agri-food-nutrition and healthcare. Omics J Integr Biol 19(5):261–275
Ramalingam A, Kudapa H, Pazhamala LT, Weckwerth W, Varshney RK (2015) Proteomics and metabolomics: two emerging areas for legume improvement. Front Plant Sci 24:6:1116
Rangan P, Furtado A, Henry RJ (2017) The transcriptome of the developing grain: a resource for understanding seed development and the molecular control of the functional and nutritional properties of wheat. BMC Genom 18(1):766
Rodrigues AS, Miguel CM (2017) The pivotal role of small non-coding RNAs in the regulation of seed development. Plant Cell Rep 36(5):653–667
Rogers K, Chen X (2013) Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 1:tpc-113
Salekdeh GH, Komatsu S (2007) Crop proteomics: aim at sustainable agriculture of tomorrow. Proteomics 7(16):2976–2996
Sato K, Yamane M, Yamaji N, Kanamori H, Tagiri A, Schwerdt JG, Fincher GB, Matsumoto T, Takeda K, Komatsuda T (2016) Alanine aminotransferase controls seed dormancy in barley. Nat Commun 7:11625
Schrick K, Mayer U, Horrichs A, Kuhnt C, Bellini C, Dangl J, Schmidt J, Jürgens G (2000) FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev 14(12):1471–1484
Souza AL, Lopez-Losano JL, Angelo PC, Souza-Neto JN, Cordeiro IB, Astolfi-Filho S, Andrade EV (2017) A proteomic approach to guarana seed and pericarp maturation. Genet Mol Res 16(3)
Sreenivasulu N (2017) Systems biology of seeds: deciphering the molecular mechanisms of seed storage, dormancy and onset of germination. Plant Cell Rep 36:633–635
Sreenivasulu N, Wobus U (2013) Seed-development programs: a systems biology-based comparison between dicots and monocots. Annu Rev Plant Biol 64:189–217
Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci 98(20):11806–11811
Sucaet Y, Wang Y, Li J, Wurtele ES (2012) MetNet Online: a novel integrated resource for plant systems biology. BMC Bioinform 13(1):267
Sudarshan GP, Kulkarni M, Akhov L, Ashe P, Shaterian H, Cloutier S, Rowland G, Wei Y, Selvaraj G (2017) QTL mapping and molecular characterization of the classical D locus controlling seed and flower color in Linum usitatissimum (flax). Sci Rep 7(1):15751
Tang G, Galili G, Zhuang X (2007) RNAi and microRNA: breakthrough technologies for the improvement of plant nutritional value and metabolic engineering. Metabolomics 3(3):357–369
Tao Z, Shen L, Gu X, Wang Y, Yu H, He Y (2017) Embryonic epigenetic reprogramming by a pioneer transcription factor in plants. Nature 551(7678):124
Venglat P, Xiang D, Wang E, Datla R (2014) Genomics of seed development: challenges and opportunities for genetic improvement of seed traits in crop plants. Biocatal Agric Biotechnol 3(1):24–30
Wan JM, Cao YJ, Wang CM, Ikehashi H (2005) Quantitative trait loci associated with seed dormancy in rice. Crop Sci 45(2):712–716
Wang H, Schauer N, Usadel B, Frasse P, Zouine M, Hernould M, Latché A, Pech JC, Fernie AR, Bouzayen M (2009) Regulatory features underlying pollination-dependent and -independent tomato fruit set revealed by transcript and primary metabolite profiling. Plant Cell 21(5):1428–1452
Wang HL, Tian CY, Wang L (2017) Germination of dimorphic seeds of Suaeda aralocaspica in response to light and salinity conditions during and after cold stratification. PeerJ 5:e3671
Watson L, Henry RJ (2005) Microarray analysis of gene expression in germinating barley embryos (Hordeum vulgare L.). Funct Integr Genom 5(3):155–162
Westoby M, Falster DS, Moles AT, Vesk PA, Wright IJ (2002) Plant ecological strategies: some leading dimensions of variation between species. Annu Rev Ecol Syst 33(1):125–159
Willmann MR, Mehalick AJ, Packer RL, Jenik PD (2011) MicroRNAs regulate the timing of embryo maturation in Arabidopsis. Plant Physiol 1:pp–110
Wurtele ES, Li L, Berleant D, Cook D, Dickerson JA, Ding J, Hofmann H, Lawrence M, Lee EK, Li J, Mentzen W (2007) Metnet: systems biology tools for arabidopsis. In: Concepts in plant metabolomics. Springer, Dordrecht, pp 145–157
Xie Q, Mayes S, Sparkes DL (2015) Carpel size, grain filling, and morphology determine individual grain weight in wheat. J Exp Bot 66(21):6715–6730
Xu M, Hu T, Zhao J, Park MY, Earley KW, Wu G, Yang L, Poethig RS (2016) Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet 12(8):e1006263
Zhai L, Xu L, Wang Y, Huang D, Yu R, Limera C, Gong Y, Liu L (2014) Genome-wide identification of embryogenesis-associated microRNAs in radish (Raphanus sativus L.) by high-throughput sequencing. Plant Mol Biol Rep 32(4):900–915
Zhang WH, Zhou Y, Dibley KE, Tyerman SD, Furbank RT, Patrick JW (2007) Nutrient loading of developing seeds. Funct Plant Biol 34(4):314–331
Zhang YC, Yu Y, Wang CY, Li ZY, Liu Q, Xu J, Liao JY, Wang XJ, Qu LH, Chen F, Xin P (2013) Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat Biotechnol 9:848
Zhou XR, Callahan DL, Shrestha P, Liu Q, Petrie JR, Singh SP (2014) Lipidomic analysis of Arabidopsis seed genetically engineered to contain DHA. Front Plant Sci 5:419
Acknowledgements
Authors acknowledged Biotechnology Information System Network (BTISNet), Department of Biotechnology (DBT), Govt. of India, New Delhi for the financial assistance. This work was also supported by Science and Engineering Research Board (SERB) New Delhi, India, in the form of “Fast Track Scheme for Young Scientist” awarded to MS (Grant no. YSS/2015/001278) and SG (Grant no. YSS/2015/00536). Author RKP is supported by fellowship from CSIR, India. Bioinformatics Centre (Sub-DIC) at G. B. Pant University of Agriculture and Technology, Pantnagar, India is also acknowledged for providing computational facilities.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Kumar, A., Pathak, R.K., Gayen, A. et al. Systems biology of seeds: decoding the secret of biochemical seed factories for nutritional security. 3 Biotech 8, 460 (2018). https://doi.org/10.1007/s13205-018-1483-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-018-1483-9