Abstract
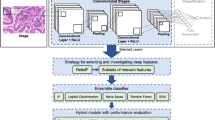
Recognition of tissues and organs is a recurrent step performed by experts during analyses of histological images. With advancement in the field of machine learning, such steps can be automated using computer vision methods. This paper presents an ensemble-based approach for improved classification of non-pathological tissues and organs in histological images using convolutional neural networks (CNNs). With limited dataset size, we relied upon transfer learning where pre-trained CNNs are re-used for new classification problems. The transfer learning was done using eleven CNN architectures upon 6000 image patches constituting training and validation subsets of a public dataset containing six cardiovascular categories. The CNN models were fine-tuned upon a much larger dataset obtained by augmenting training subset to obtain agreeable performance on validation subset. Lastly, we created various ensembles of trained classifiers and evaluate them on testing subset of 7500 patches. The best ensemble classifier gives, precision, recall, and accuracy of 0.876, 0.869 and 0.869, respectively upon test images. With an overall F1-score of 0.870, our ensemble-based approach outperforms previous approaches with single fine-tuned CNN, CNN trained from scratch, and traditional machine learning by 0.019, 0.064 and 0.183, respectively. Ensemble approach can perform better than individual classifier-based ones, provided the constituent classifiers are chosen wisely. The empirical choice of classifiers reinforces the intuition that models which are newer and outperformed in their native domain are more likely to outperform in transferred-domain, since the best ensemble dominantly consists of more lately proposed and better architectures.




Similar content being viewed by others
Data availability
The dataset used in the work is a public dataset.
References
Webster JD, Dunstan RW (2014) Whole-slide imaging and automated image analysis: considerations and opportunities in the practice of pathology. Vet Pathol 51(1):211–223. https://doi.org/10.1177/0300985813503570
Mccann T et al (2015) Automated histology analysis. IEEE Signal Process Mag 32(1):78–87
Jansen I et al (2018) Histopathology: ditch the slides, because digital and 3D are on show. World J Urol 6(4):549–555. https://doi.org/10.1007/s00345-018-2202-1
Madabhushi A, Lee G (2016) Image analysis and machine learning in digital pathology: challenges and opportunities. Med Image Anal 33:170–175. https://doi.org/10.1016/j.media.2016.06.037
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436–444. https://doi.org/10.1038/nature14539
Goodfellow I, Bengio Y, Courville A (2016) Deep learning. MIT Press, Cambridge
Litjens G et al (2017) A survey on deep learning in medical image analysis. Med Image Anal 42(1995):60–88. https://doi.org/10.1016/j.media.2017.07.005
Tiwari P et al (2018) Detection of subtype blood cells using deep learning. Cogn Syst Res 52:1036–1044. https://doi.org/10.1016/j.cogsys.2018.08.022
Harangi B (2018) Skin lesion classification with ensembles of deep convolutional neural networks. J Biomed Inform 86:25–32. https://doi.org/10.1016/j.jbi.2018.08.006
Sharma H, Zerbe N, Klempert I, Hellwich O, Hufnagl P (2017) Deep convolutional neural networks for automatic classification of gastric carcinoma using whole slide images in digital histopathology. Comput Med Imaging Graph. https://doi.org/10.1016/j.compmedimag.2017.06.001
Saha M, Chakraborty C, Racoceanu D (2018) Efficient deep learning model for mitosis detection using breast histopathology images. Comput Med Imaging Graph 64:29–40. https://doi.org/10.1016/j.compmedimag.2017.12.001
Almubarak HA et al (2017) Convolutional neural network based localized classification of uterine cervical cancer digital histology images. Procedia Comput Sci 114:281–287. https://doi.org/10.1016/j.procs.2017.09.044
Das DK, Bose S, Maiti AK, Mitra B, Mukherjee G, Dutta PK (2018) Automatic identification of clinically relevant regions from oral tissue histological images for oral squamous cell carcinoma diagnosis. Tissue Cell 53(June):111–119. https://doi.org/10.1016/j.tice.2018.06.004
Mazo C, Bernal J, Trujillo M, Alegre E (2018) Transfer learning for classification of cardiovascular tissues in histological images. Comput Methods Programs Biomed 165:69–76. https://doi.org/10.1016/j.cmpb.2018.08.006
Mazo C, Alegre E, Trujillo M (2017) Classification of cardiovascular tissues using LBP based descriptors and a cascade SVM. Comput Methods Programs Biomed 147:1–10. https://doi.org/10.1016/j.cmpb.2017.06.003
Zeiler MD, Fergus R (2014) Visualizing and understanding convolutional networks. In: European conference on computer vision, vol 12. Springer, Cham, pp 818–833. https://doi.org/10.1016/j.ancr.2017.02.001.
Yosinski J, Clune J, Bengio Y, Lipson H (2014) How transferable are features in deep neural networks? Adv Neural Inf Process Syst 27:3320–3328
Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L (2009) ImageNet: a large-scale hierarchical image database. In: Conference on computer vision and pattern recognition (CVPR 2009). https://doi.org/10.1167/9.8.1037
Chollet F (2015) Keras: deep learning library for Theano and TensorFlow. https://keras.io
Abadi M et al (2016) TensorFlow: large-scale machine learning on heterogeneous distributed systems. http://www.tensorflow.org. http://arxiv.org/abs/1603.04467
Pedregosa F et al (2011) Scikit-learn: machine learning in Python. J Mach Learn Res 12:2825–2830
Howard AG et al (2017) MobileNets: efficient convolutional neural networks for mobile vision applications. http://arxiv.org/abs/1704.04861
Simonyan K, Zisserman A (2015) Very deep convolutional networks for large-scale image recognition. In: International conference on learning representations 2015, pp 1–14. http://arxiv.org/abs/1409.1556
Huang G, Liu Z, van der Maaten L, Weinberger KQ (2016) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition (CVPR), 2017, pp 4700–4708. https://doi.org/10.1109/CVPR.2017.243
Zoph B, Vasudevan V, Shlens J, Le QV (2018) Learning transferable architectures for scalable image recognition. In: Proceedings of the IEEE computer society conference on computer vision and pattern recognition, pp 8697–8710. https://doi.org/10.1109/CVPR.2018.00907
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of IEEE computer society conference on computer vision and pattern recognition, December 2016, pp 770–778. https://doi.org/10.1109/CVPR.2016.90
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: Proceedings of IEEE computer society conference on computer vision and pattern recognition, pp 2818–2826. https://doi.org/10.1002/2014GB005021.
Chollet F (2017) Xception: Deep learning with depthwise separable convolutions. In: Proceedings of the 30th IEEE conference on computer vision and pattern recognition, CVPR 2017, January 2017, pp 1800–1807. https://doi.org/10.1109/CVPR.2017.195
Szegedy C, Ioffe S, Vanhoucke V (2016) Inception-v4, Inception-ResNet and the impact of residual connections on learning. In: AAAI conference on artificial intelligence, p 12. https://doi.org/10.1016/j.patrec.2014.01.008.
Gonzalez RC, Woods RE (2014) Digital image processing. Addison-Wesley, Reading
Dunham MH (2003) Data mining: introductory and advanced topics. Pearson Education India, Delhi
Lin M, Chen Q, Yan S (2013) Network in network. http://arxiv.org/abs/1312.4400v3, pp 1–10. https://doi.org/10.1109/ASRU.2015.7404828.
Kingma DP, Adam JB (2015) A method for stochastic optimization. In: International conference on learning representations, pp 1–15. http://arxiv.org/abs/1412.6980.
Chollet F (2018) Deep learning with Python. Manning Publications, Shelter Island
Polikar R (2006) Ensemble based systems in decision making. IEEE Circuits Syst Mag 61:21–45
Rokach L (2010) Ensemble-based classifiers. Artif Intell Rev 33(1–2):1–39. https://doi.org/10.1007/s10462-009-9124-7
Leo B (1996) Bagging predictors. Mach Learn 24(2):123–140. https://doi.org/10.1007/BF00058655
Cutler A, Cutler DR, Stevens JR (2012) Random forests. In: Ensemble machine learning: methods and applications. Springer, Boston, pp 157–175. https://doi.org/10.1007/9781441993267_5
Opitz DW, Shavlik JW (1996) Generating accurate and diverse members of a neural-network ensemble. Adv Neural Inf Process Syst 8:535–541
Wen G, Hou Z, Li H, Li D, Jiang L, Xun E (2017) Ensemble of deep neural networks with probability-based fusion for facial expression recognition. Cognit Comput 9(5):597–610. https://doi.org/10.1007/s12559-017-9472-6
Boser BE, Guyon IM, Vapnik VN (1999) A training algorithm for optimal margin classifiers. In: Proceedings of the fifth annual workshop on Computational learning theory, pp 144–152. https://doi.org/10.1145/130385.130401.
Kuncheva LI, Whitaker CJ (2003) Measures of diversity in classifier ensembles and their relationship with the ensemble accuracy. Mach Learn 51:181–207
Yule GU (1912) On the methods of measuring association between two attributes. J R Stat Soc 75(6):579–652
Wu R, Yan S, Shan Y, Dang Q, Sun G (2015) Deep image: scaling up image recognition. http://arxiv.org/abs/1501.02876
Wittman T (2005) Mathematical techniques for image interpolation. http://public-digital-library.googlecode.com/svn/trunk/DSP/ImageResampling/MathematicalTechniquesforImageInterpolation.pdf
Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D (2017) Grad-CAM: visual explanations from deep networks via gradient-based localization. In: Proceedings of IEEE international on conference on computer vision, October 2017, pp 618–626. https://doi.org/10.1109/ICCV.2017.74
Kotikalapudi R et al (2017) keras-vis. https://github.com/raghakot/keras-vis
Funding
None.
Author information
Authors and Affiliations
Contributions
The work is contributed by a single author.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent for publication
The publisher has the author’s permission to publish the relevant work.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
No animals or humans were involved in this research.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mittal, S. Ensemble of transfer learnt classifiers for recognition of cardiovascular tissues from histological images. Phys Eng Sci Med 44, 655–665 (2021). https://doi.org/10.1007/s13246-021-01013-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13246-021-01013-2