Abstract
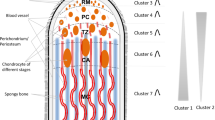
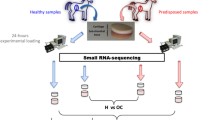
Deer antlers are the only mammalian appendages subject to an annual cycle of epimorphic regeneration. Within the rapid-growth stage, they display the fastest cartilage development in the animal kingdom. To identify microRNA (miRNA) profiling in red deer (Cervus elaphus) antler cartilage, we applied deep sequencing technology to a small RNA library constructed from pooled cartilage (three antlers from three individuals). We generated 9,520,645 mappable reads with a size distribution of between 15 and 30 nucleotides (miRNAs of 18 nucleotides were the most abundant group: 31 %). Bioinformatics data mining revealed 399 miRNAs in antler cartilage, of which 345 were highly conserved and expressed in 25 other mammals, including the cattle (Bos taurus) and in humans (Homo sapiens). The remaining 54 miRNAs we identified were novel and likely to be antler-cartilage specific, but were expressed at low levels. The identification of these known and newly identified miRNAs in antler cartilage significantly enhances our understanding of the miRNA profiling of regenerating antler cartilage. Further studies are necessary to better understand miRNAs-regulated antlerogenesis.


Similar content being viewed by others
References
Allen SP, Maden M, Price JS (2002) A role for retinoic acid in regulating the regeneration of deer antlers. Dev Biol 251:409–423
Ambady S, Wu Z, Dominko T (2012) Identification of novel MicroRNAs in Xenopus laevis metaphase II arrested eggs. Genesis 50:286–299
Ambros V (2001) microRNAs: tiny regulators with great potential. Cell 107:823–826
Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M et al (2003) A uniform system for microRNA annotation. RNA 9:277–279
Banks JW, Newbrey WJ (1983a) Light microscopic studies of the ossification process in developing antlers. In: Brown RD (ed) Antler Development in cervidea: a proceedings of the first international symposium of the Caesar Kleberg Wildlife Research Institute, College of Agriculture, Texas A&I University, Kingsville, Texas. Kingsville: Caesar Kleberg Wildlife Research Institute, pp 231–260
Banks JW, Newbry WJ (1983) Antler development as a unique modification of mammalian endochondral ossification. In: Banks RD (ed) Antler developments in cervidae. Caesar Kleburg Wildlife Research Institute, Kingsville, pp 279–306
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Cai Y, Yu X, Zhou Q, Yu C, Hu H, Liu J, Lin H, Yang J, Zhang B, Cui P et al (2010) Novel microRNAs in silkworm (Bombyx mori). Funct Integr Genomics 10:405–415
Cai EH, Gao YX, Wei ZZ, Chen WY, Yu P, Li K (2012) Serum miR-21 expression in human esophageal squamous cell carcinomas. Asian Pac J Cancer Prev 13:1563–1567
Camarillo C, Swerdel M, Hart RP (2011) Comparison of microarray and quantitative real-time PCR methods for measuring MicroRNA levels in MSC cultures. Methods Mol Biol 698:419–429
Caponi S, Funel N, Frampton AE, Mosca F, Santarpia L, Van der Velde AG, Jiao LR, De Lio N, Falcone A, Kazemier G et al (2012) The good, the bad and the ugly: a tale of miR-101, miR-21 and miR-155 in pancreatic intraductal papillary mucinous neoplasms. Ann Oncol 24:734–741
Chapman DI (1975) Antlers-bones of contention. Mamm Rev 5:121–172
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR et al (2005) Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33:e179–e187
Chen X, Li Q, Wang J, Guo X, Jiang X, Ren Z, Weng C, Sun G, Wang X, Liu Y et al (2009) Identification and characterization of novel amphioxus microRNAs by Solexa sequencing. Genome Biol 10:R78–R90
Chen YH, Wang SQ, Wu XL, Shen M, Chen ZG, Chen XG, Liu YX, Zhu XL, Guo F, Duan XZ et al (2011) Characterization of microRNAs expression profiling in one group of Chinese urothelial cell carcinoma identified by Solexa sequencing. Urol Oncol 31:219–227
Chen C, Deng B, Qiao M, Zheng R, Chai J, Ding Y, Peng J, Jiang S (2012) Solexa sequencing identification of conserved and novel microRNAs in backfat of Large White and Chinese Meishan pigs. PLoS One 7:e31426–e31435
Danks J, Dacke C, Flik G, Gay C (1999) Calcium metabolism: comparative endocrinology. Bio Scientifica Ltd, Bristol, pp 131–138
de Crombrugghe B, Lefebvre V, Behringer RR, Bi W, Murakami S, Huang W (2000) Transcriptional mechanisms of chondrocyte differentiation. Matrix Biol 19:389–394
Dippold RP, Vadigepalli R, Gonye GE, Hoek JB (2012) Chronic ethanol feeding enhances miR-21 induction during liver regeneration while inhibiting proliferation in rats. Am J Physiol Gastrointest Liver Physiol 303:G733–G743
Drayton RM (2012) The role of microRNA in the response to cisplatin treatment. Biochem Soc Trans 40:821–825
Faragalla H, Youssef YM, Scorilas A, Khalil B, White NM, Mejia-Guerrero S, Khella H, Jewett MA, Evans A, Lichner Z et al (2012) The clinical utility of miR-21 as a diagnostic and prognostic marker for renal cell carcinoma. J Mol Diagn 14:385–392
Faucheux C, Nesbitt SA, Horton MA, Price JS (2001) Cells in regenerating deer antler cartilage provide a microenvironment supports osteoclast differentiation. J Exp Biol 204:443–455
Faucheux C, Horton MA, Price JS (2002) Nuclear localization of type I parathyroid hormone/parathyroid hormone-related protein receptors in deer antler osteoclasts: evidence for parathyroid hormone-related protein and receptor activator of NF-κB-dependent effects on osteoclast formation in regenerating mammalian bone. J Bone Miner Res 17:455–464
Faucheux C, Nicholls BM, Allen S, Danks JA, Horton MA, Price JS (2004) Parathyroid hormone-related peptide may play a role in deer antler regeneration. Dev Dyn 231:88–97
Friedman RC, Farh KK, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19:92–105
Goss RJ (1970) Problems of antlerogenesis. Clin Orthopaed 69:227–238
Goss RJ (1983) Deer antlers: regeneration, function and evolution. Academic Press, New York
Gu Z, Eleswarapu S, Jiang H (2007) Identification and characterization of microRNAs from the bovine adipose tissue and mammary gland. FEBS Lett 581:981–988
Hannon GJ (2002) RNA interference. Nature 418:244–251
Hermansen SK, Dahlrot RH, Nielsen BS, Hansen S, Kristensen BW (2012) MiR-21 expression in the tumor cell compartment holds unfavorable prognostic value in gliomas. J Neurooncol 111:71–81
Hu W, Meng X, Lu T, Wu L, Li T, Li M, Tian Y (2013) MicroRNA-1 inhibits the proliferation of Chinese sika deer-derived cartilage cells by binding to the 3′-untranslated region of IGF-1. Mol Med Rep 8:523–528
Hu W, Li T, Wu L, Li M, Meng X (2014) Identification of microRNA-18a as a novel regulator of the insulin-like growth factor-1 in the proliferation and regeneration of deer antler. Biotechnol Lett 36:703–710
Huang J, Ju Z, Li Q, Hou Q, Wang C, Li J, Li R, Wang L, Sun T, Hang S et al (2011) Solexa sequencing of novel and differentially expressed microRNAs in testicular and ovarian tissues in Holstein cattle. Int J Biol Sci 7:1016–1026
Ji Z, Wang G, Xie Z, Zhang C, Wang J (2012) Identification and characterization of microRNA in the dairy goat (Capra hircus) mammary gland by Solexa deep-sequencing technology. Mol Biol Rep 39:9361–9371
Jiang J, Lee EJ, Gusev Y, Schmittgen TD (2005) Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucl Acids Res 33:5394–5403
Kierdorf U, Li C, Price JS (2009) Improbable appendages: deer antler renewal as a unique case of mammalian regeneration. Semin Cell Dev Biol 20:535–542
Kobayashi T, Lu J, Cobb BS, Rodda SJ, McMahon AP, Schipani E, Merkenschlager M, Kronenberg HM (2008) Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc Natl Acad Sci USA 105:1949–1954
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854
Legeai F, Rizk G, Walsh T, Edwards O, Gordon K, Lavenier D, Leterme N, Méreau A, Nicolas J, Tagu D et al (2010) Bioinformatic prediction, deep sequencing of microRNAs and expression analysis during phenotypic plasticity in the pea aphid, Acyrthosiphon pisum. BMC Genom 11:281–289
Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB (2003) Prediction of mammalian microRNA targets. Cell 115:787–798
Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20
Li C (2012) Deer antler regeneration: a stem cell-based epimorphic process. Birth Defects Res C Embryo Today 96:51–62
Li C, Clark DE, Lord EA, Stanton JA, Suttie JM (2002) Sampling technique to discriminate the different tissue layers of growing antler tips for gene discovery. Anat Rec 268:125–130
Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433:769–773
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA et al (2005) MicroRNA expression profiles classify human cancers. Nature 435:834–838
Nakamura Y, Inloes JB, Katagiri T, Kobayashi T (2011) Chondrocyte-specific microRNA-140 regulates endochondral bone development and targets Dnpep to modulate bone morphogenetic protein signaling. Mol Cell Biol 31:3019–3028
Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Müller P et al (2000) Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408:86–89
Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W (2005) Inhibition of translational initiation by let-7 microRNA in human cells. Science 309:1573–1576
Price JS, Oyajobi BO, Oreffo RO, Russell RG (1994) Cells cultured from the growing tip of red deer antler express alkaline phosphatase and proliferate in response to insulin-like growth factor-I. J Endocrinol 143:R9–R16
Price JS, Oyajobi BO, Nalin AM, Frazer A, Russell RG, Sandell LJ (1996) Chondrogenesis in the regenerating antler tip in red deer: expression of collagen types I, IIA, IIB, and X demonstrated by in situ nucleic acid hybridization and immunocytochemistry. Dev Dyn 205:332–347
Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Genes Dev 16:1616–1626
Sayed D, Abdellatif M (2011) MicroRNAs in development and disease. Physiol Rev 91:827–887
Schmittgen TD, Jiang J, Liu Q, Yang L (2004) A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res 32:e43–e52
Sdassi N, Silveri L, Laubier J, Tilly G, Costa J, Layani S, Vilotte JL, Le Provost F (2009) Identification and characterization of new miRNAs cloned from normal mouse mammary gland. BMC Genom 10:149–158
Sun J, Zhong N, Li Q, Min Z, Zhao W, Sun Q, Tian L, Yu H, Shi Q, Zhang F et al (2011) MicroRNAs of rat articular cartilage at different developmental stages identified by Solexa sequencing. Osteoarthr Cartil 19:1237–1245
Szuwart T, Kierdorf H, Kierdorf U, Clemen G (2002) Histochemical and ultrastructural studies of cartilage resorption and acid phosphatase activity during antler growth in fallow deer (Dama dama). Anat Rec 268:66–72
Vaz C, Ahmad HM, Sharma P, Gupta R, Kumar L, Kulshreshtha R, Bhattacharya A (2010) Analysis of microRNA transcriptome by deep sequencing of small RNA libraries of peripheral blood. BMC Genom 11:288–305
Wei Z, Liu X, Feng T, Chang Y (2011) Novel and conserved microRNAs in Dalian purple urchin (Strongylocentrotus nudus) identified by next generation sequencing. Int J Biol Sci 7:180–192
Wheeler BM, Heimberg AM, Moy VN, Sperling EA, Holstein TW, Heber S, Peterson KJ (2009) The deep evolution of metazoan microRNAs. Evol Dev 11:50–68
Yekta S, Shih IH, Bartel DP (2004) MicroRNA-directed cleavage of HOXB8 mRNA. Science 304:594–596
Zhang BH, Wang QL, Pan XP (2007) MicroRNAs and their regulatory roles in animals and plants. J Cell Physiol 210:279–289
Zhao Y, Yao B, Zhang M, Wang S, Zhang H, Xiao W (2013) Comparative analysis of differentially expressed genes in Sika deer antler at different stages. Mol Biol Rep 40:1665–1676
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415
Acknowledgments
We thank Mr. Shouzhuang YANG at the Qinghuangdao Safari Park for his help and technical support.
Conflict of interest
The authors declare that there is no conflict of interest.
Ethical Statement
This study was subject to approval by the Animal Ethics Committee of the Northeast Forestry University (AEC-NEFU; Permit Number: 2012-0016).
Funding
This work was funded by the Fundamental Research Funds for the Central Universities (No. 2572014EA05-01 to DZ) and the Program for New Century Excellent Talents in University (No. NCET-11-0609 to DZ).
Author information
Authors and Affiliations
Corresponding author
Additional information
Yanxia Chen and Xuedong Liu have equally contributed to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, Y., Liu, X., Yang, X. et al. Deep sequencing identifies conserved and novel microRNAs from antlers cartilage of Chinese red deer (Cervus elaphus). Genes Genom 37, 419–427 (2015). https://doi.org/10.1007/s13258-015-0270-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-015-0270-9