Abstract
Glyphosate is a broad-spectrum systemic herbicide used to kill weeds, especially annual broadleaf weeds and grasses known to compete with commercial crops grown around the globe. However, weeds evolve and develop resistance to glyphosate. Until recently, no case of glyphosate resistance had been detected in France. Glyphosate resistance was indeed recently recorded in a Lolium rigidum weed population from a vineyard in the South of France. Here, we studied the mechanisms of this resistance case. Seed samples of L. rigidum were collected from the vineyard where resistance had been detected, as well as from a nearby area that had no known history of exposure to glyphosate. We studied the effect of retention of glyphosate spray, shikimic acid accumulation, glyphosate absorption and translocation, glyphosate metabolism, and the sequence of the enzyme that glyphosate targets in plants, 5-enolpyruvylshikimate-3-phosphate synthase. Our results show that glyphosate absorption decreased by 30 % in the resistant L. rigidum weed. In addition, glyphosate translocation out of the treated leaves was reduced by 52 %. Finally, the resistant biotype had a serine amino acid substitution at position 106 of the predicted protein, instead of the proline amino acid present in the susceptible population. Our results suggest that the resistant population of L. rigidum presents three different mechanisms of resistance to glyphosate, namely reduced absorption, reduced mobility in the plants, and a mutation in the gene coding for the enzyme targeted by glyphosate.
Similar content being viewed by others
1 Introduction
Glyphosate is a broad-spectrum herbicide, acting exclusively via foliar uptake. It has been extensively used to control plants in various crops, tillage systems, and non-agricultural areas (Franz et al. 1997). Glyphosate kills plants by inhibiting the enzyme 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) (EC 2.5.1.19), which catalyzes the conversion of shikimate-3-phosphate and phosphoenolpyruvate to 5-enolpyruvylshikimate-3-phosphate and inorganic phosphate in the shikimic acid pathway (Geiger and Fuchs 2002). Inhibiting this enzyme prevents the biosynthesis of aromatic amino acids: phenylalanine, tyrosine, and tryptophan.
Glyphosate application on large weed-infested areas has led to the emergence of herbicide-resistant biotypes, typically after 10 years of repeated applications (Owen 2001). Worldwide, resistance to glyphosate has been observed in 30 two weed species and subspecies, six of them (i.e., Conyza bonariensis (L.) Cronquist, Conyza canadensis (L.) Cronquist, Conyza sumatrensis (Retz.) E. Walker, Lolium multiflorum Lam., Lolium perenne L., and Lolium rigidum Gaudin occurring in Europe) (Heap 2015). In some parts of the world such as the USA and Brazil, resistance to glyphosate is mostly associated with genetically engineered crops that are tolerant to this herbicide (Powles and Yu 2010). Apart from a recent, not fully characterized, case (Collavo and Sattin 2014), all resistance cases recorded so far in Europe were observed in perennial crops or in non-crop areas (Heap 2015). The first case of glyphosate resistance in France was detected in a vineyard and concerned a L. rigidum population (Favier and Gauvrit 2007), and only more recently another glyphosate-resistant biotype of C. sumatrensis has been detected (Heap 2015).
Resistance to glyphosate has been ascribed to the following factors: (a) reduced foliar retention of the herbicide (González-Torralva et al. 2010), (b) reduced absorption of the herbicide (Michitte et al. 2007), (c) reduced translocation of the herbicide probably due to sequestration into the vacuole (Ge et al. 2010), (d) mutations in DNA coding sequences leading to an altered resistant form of EPSPS (Powles and Yu 2010; González-Torralva et al. 2012), (e) overexpression of EPSPS through gene amplification (Gaines et al. 2010), and (f) metabolization of glyphosate into non-toxic compounds (De Carvalho et al. 2012). To date, the mechanisms involved in the French resistance case are not known, so the purpose of this study was to elucidate them.
2 Material and methods
2.1 Plant material and growing conditions
Seeds of L. rigidum Gaudin were collected in the summer of 2009 at Saint Pons la Calm (South of France, lat. 44° 05′ N, long. 4° 33′ E). A sample was collected in a vineyard where 1080 to 1800 g ha−1 glyphosate (roundup, 360 g ae L−1 as isopropylamine salt) had been applied annually for at least 10 years and where observations had been made that glyphosate failed to control L. rigidum. The other sample was obtained from a field nearby that had never received glyphosate treatments.
All seeds were germinated in Petri dishes holding filter paper moistened with distilled water and placed in growth chambers at 28/18 °C (day/night) with a photoperiod of 16 h, 850 μmol m−2 s−1 photosynthetic photon flux, and 80 % relative humidity. Seedlings of both resistant and susceptible populations from each species were transplanted into pots (three plants per pot) containing sand/peat in a 1:2 (v/v) ratio and placed in a growth chamber under the conditions described.
2.2 Dose–response assays
Glyphosate applications were made at the 3- to 4-leaf stage, with a laboratory spray chamber (SBS-060 DeVries Manufacturing) equipped with 8002 flat fan nozzles delivering 200 L ha−1 at 200 kPa at a height of 50 cm. The following glyphosate (Roundup Energy SL, 450 g ae L−1 as isopropylamine salt, Monsanto) rates were used: 0, 25, 50, 75, 100, 150, 200, 250, 300, 350, 400, 450, 500, 550, and 600 g ae ha−1. The experiment was arranged in a completely randomized design using four replicates (each replicate with three plants) per rate. Plants were harvested 21 days after treatment and immediately weighed to determine fresh weight. Data were expressed as half-effective doses (ED50, the glyphosate rate needed to reduce fresh weight by 50 %) and compared to the non-treated control plants of both biotypes.
2.3 Spray retention assays
L. rigidum plants at the 3- to 4-leaf stage were treated with a solution containing 100 mg L−1 Na-fluorescein and glyphosate at a concentration corresponding to 300 g ae ha−1 applied in 200 L ha−1 using the laboratory spray chamber and conditions previously described. After the spray had dried on the foliage (approximately 20 to 25 min), the plants were cut off at ground level and immersed in 50 mL of 5 mM NaOH for 30 s. The rinsate of each plant was then filtered and kept in vials to measure immediately its fluorescein concentration in a Hitachi F-2500 fluorescence spectrophotometer at 490exc/510em nm. Plants were then placed in a heater (J. P. Selecta S.A.) at 60 °C for 48 h and dry matter weighed. The experiment was arranged in a completely randomized design with four replicates (three plants each) per species. The results were expressed as milliliters spraying solution per gram dry matter (González-Torralva et al. 2010).
2.4 Shikimic acid accumulation
Plants at the 3- to 4-leaf stage were treated with glyphosate at 300 g ae ha−1 with the laboratory spray chamber and same conditions as described above. At 12, 24, 48, 72, and 96 h after treatment, 50 mg of plant tissue was harvested and placed in a vial containing 1 mL of 1 M HCl and then immediately frozen in liquid nitrogen. Shikimic acid accumulation was determined according to Singh and Shaner (1998). Sample absorbance was measured with a Beckman DU-640 spectrophotometer at 380 nm. Net shikimic acid accumulation was deduced from the difference between treated and non-treated plants in each population. The test was performed in triplicate on five treated and five non-treated plants per species, and the results were expressed as micrograms per gram fresh weight. The rate of shikimic acid accumulation (μg g−1 fresh weight h−1) was measured between 12 and 96 h after treatment.
2.5 Absorption and translocation of 14C-glyphosate
In order to have the wetting agents and additives needed for absorption, 14C-glyphosate (American Radiolabeled Chemicals, Inc., Saint Louis, MO, USA) was added to the commercial herbicide to prepare a solution with a specific activity of 0.834 kBq μL−1. The final glyphosate concentration corresponded to 300 g ae ha−1 applied in 200 L ha−1. One 1-μL droplet was deposited by means of a micropipette (LabMate + HTL) onto the adaxial surface of the second leaf of plants at the 3-leaf stage (0.834 kBq/plant). Preliminary assays with the two populations studied here had revealed that glyphosate absorption levelled-off at 96 h after droplet application (results not shown). The treated leaf from each plant was carefully washed with 3 mL of water: acetone (1:1 v/v) solution to remove the unabsorbed 14C-glyphosate. The rinsate was mixed with 2 mL of scintillation cocktail and analyzed by liquid scintillation spectrometry (LSS) on a Beckman LS 6500 scintillation counter. The remainder of the plant was carefully removed from the pot, and its roots were carefully washed with distilled water. The plant was divided into treated leaf, remaining shoot tissue, and roots. The plant parts thus obtained were dried at 60 °C for 96 h and combusted in a Packard Tri Carb 307 biological sample oxidizer. Evolved 14CO2 was trapped and counted by LSS in a 18-mL mixture of Carbo-Sorb E and Permafluor E+ (1:1 v/v) (Perkin-Elmer, Packard Bioscience BV). The proportion of absorbed herbicide was expressed as [kBq in combusted tissue / (kBq in combusted tissue + kBq in leaf washes)] × 100. There were three replicates (each one with three plants), and the experiment was arranged in a completely randomized design. The amount of radiolabel deposited was checked by washing a treated leaf excised immediately after deposition (three replicates). The mean radioactivity recoveries (SE) were 92 (6.3) % and 94 (2.7) % for resistant and susceptible L. rigidum, respectively.
2.6 14C-glyphosate visualization
14C-glyphosate translocation was visualized in plants from the resistant and susceptible populations of L. rigidum using a phosphor imager (Cyclone, Perkin-Elmer). Plants were treated and collected in the same way as described in the absorption and translocation assays. The whole plants were gently rinsed, pressed, and then let to dry at room temperature during 4 days. Then, the dried plants were placed adjacent to a 25 cm × 12.5 cm phosphor storage film for 14 h and scanned for radiolabel distribution on a phosphor imager. The experiment was carried out with three plants per each population (resistant and susceptible).
2.7 Metabolism study
L. rigidum plants at the 3- to 4-leaf stage were treated at a glyphosate rate of 300 g ae ha−1 as described in the dose-response assays section, and other plants were kept without treatment as non-treated controls. At 96 h after treatment, following the methodology described by Rojano-Delgado et al. (2010), glyphosate and its metabolites, i.e., aminomethylphosphonic acid (AMPA), glyoxylate, sarcosine, and formaldehyde, were determined by reversed polarity capillary electrophoresis using a 3D Capillary Electrophoresis Agilent G1600A instrument equipped with a diode array detector (DAD, wavelength range 190–600 nm). Glyphosate, AMPA, sarcosine, formaldehyde, and glyoxylate were used as standards. Leaf tissues were washed with distilled water, flash-frozen in liquid nitrogen, and stored at −40 °C until use. The aqueous background electrolyte consisted of 10 mM potassium phthalate, 0.5 mM hexadecyltrimethylammonium bromide, and 10 % acetonitrile at pH 7.5. The calibration equations were established from non-treated plants and known concentrations of glyphosate and its metabolites, which were determined from their peak areas in the electropherogram. The average value for the content of glyoxylate naturally produced by the plant was subtracted from the average of the content of each population. The experiment was arranged in a completely randomized design with four replications per biotype.
2.8 EPSPS gene sequencing
Total RNA was isolated from leaves using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. RNA was then treated with TURBO DNase (RNase-Free; Ambion, Warrington, UK) to eliminate any DNA contamination and stored at −80 °C. cDNA synthesis was carried out from 2 μg of total RNA using a M-MLV (Moloney Murine Leukemia Virus) Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) in combination with oligo (dT)12-18 and random nonamers (Amersham Biosciences, Amersham, UK) according to the manufacturer’s instructions. To amplify the EPSPS gene, primers previously designed by Perez-Jones et al. (2007) (forward: 5′ AGCTGTAGTCGTTGGCTGTG 3′; reverse: 5′ GCCAAGAAATAGCTCGCACT 3′) were used. These primers expand a 543-bp fragment of the EPSPS gene that contains the mutation site described as conferring resistance to glyphosate in Lolium spp. The PCR reactions were carried out using cDNA from 50 ng of total RNA, 1.5 mM MgCl2, 0.2 mMdNTP, 0.2 μM of each primer, 1× buffer, and 0.625 units of a 100:1 enzyme mixture of non-proofreading (Thermusthermophilus) and proofreading (Pyrococcusfuriosus) polymerases (BIOTOOLS, Madrid, Spain) in a final volume of 25 μL. All PCR reactions were in duplicate and cycling conditions were: 94 °C 3 min, 35 cycles of 94 °C 30 s, 55 °C 30 s and 72 °C 1 min; and a final extension cycle of 72 °C 10 min. An aliquot of the PCR product was loaded in a 1 % agarose gel to check the correct band amplification. The rest of the PCR product was then purified using ExoSAP-IT® for PCR Product Clean-Up (USB, Ohio, USA) as indicated by the manufacturers. Five purified PCR products per biotype were sequenced (STAB VIDA, Caparica, Portugal). Finally, the EPSPS DNA and the predicted peptide sequences were searched through the Gen-Bank database using the program BLAST (basic local alignment search tool) (Altschul et al. 1990) on the website (http://www.ncbi.nlm.nih.gov/BLAST/).
2.9 Statistical analyses
Data obtained in the dose response assays were pooled and fitted to a non-linear, log-logistic regression equation:
where Y is the above ground fresh weight expressed as percentage of the non-treated control, c and d are the coefficients corresponding to the lower and upper asymptotes, b is the slope of the line, ED50 is the herbicide rate at the inflection point halfway between the upper and lower asymptotes (that is the herbicide rate inhibiting plant growth of each species by 50 % with respect to the non-treated control), and x (independent variable) is the herbicide rate. Regression analysis was conducted using the Sigmaplot 10.0 statistical software.
Data obtained in spray retention, shikimic acid accumulation, absorption–translocation of 14C-glyphosate, and metabolism assays were submitted to analysis of variance (ANOVA). When necessary, data expressed as percentages were arcsin normalized and the Tukey HSD test at the 5 % probability was used to separate means. Statistical analyses were conducted using the Statistix (version. 8.0) (Analytical Software, USA) software.
3 Results and discussion
3.1 Dose–response assays and spray retention
Table 1 shows that ED50 (the glyphosate rate needed to reduce fresh weight by 50 %) was significantly higher for the resistant population of L. rigidum as compared to its susceptible counterpart. The resistance factor [ED50 (resistant)/ED50 (susceptible)] was 5.8, which is similar to those observed in other glyphosate-resistant L. rigidum biotypes (Preston et al. 2009).
Spray retention by L. rigidum plants was 1.62 (0.747) and 1.61 (0.638) mL g−1 dry weight for the resistant and susceptible populations, respectively. It can be presumed that herbicide retention is not responsible for the observed resistance. It should be noted that resistance due to reduced spray retention, although described, is rare (Michitte et al. 2007; Nandula et al. 2008).
3.2 Shikimic acid accumulation
There was no significant difference in shikimic acid accumulation between the resistant and susceptible populations at 12 h after treatment (not shown). At 96 h after treatment, there was a 6-fold increase in shikimic acid accumulation in the susceptible population of L. rigidum, as compared to the resistant population (Table 1). The accumulation of shikimic acid in plant tissues is widely accepted to reflect the inhibition of EPSPS as a result of the application of glyphosate; the higher the accumulation of shikimic acid, the greater the toxicity of glyphosate to the plants. The lower increase in shikimic acid in the resistant L. rigidum population, as compared to the susceptible population, was consistent with its resistance level.
3.3 Absorption, translocation, and visualization of 14C-glyphosate
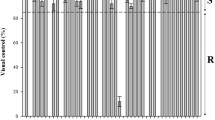
At 96 h after treatment, 60 to 86 % of applied 14C-glyphosate was absorbed (Table 1). The uptake difference between the two L. rigidum populations was significant and may play a part in the observed resistance (Fig. 1).
There were also marked differences regarding the amount of 14C-glyphosate translocated from the treated leaf to the rest of the plant (Table 1). It was about halved in the resistant population as compared to the susceptible population. In addition, translocation to the roots was more than thrice decreased. Differences in 14C-glyphosate translocation in the L. rigidum populations were also visualized by phosphor imaging (Fig. 2b). Overall, glyphosate translocation from the treated leaf at 96 h after treatment was greater and more marked in the susceptible population than in the resistant population. This visual difference was consistent with the quantified differences in the absorption and translocation of glyphosate, as well as with the differences in shikimic acid accumulation between the resistant and susceptible populations.
In the resistant L. rigidum population, reduced translocation of glyphosate to shoots and roots was clearly demonstrated. The shoots and roots are home to meristematic zones, which are sensitive to glyphosate action. Therefore, the ability to limit glyphosate translocation towards meristematic tissues may confer resistance to the plants (Shaner 2009; Yu et al. 2009).
3.4 Glyphosate metabolism
At 96 h after treatment, glyphosate was metabolized to a low extent (<15 % of absorbed) (Table 2). There were significant differences in the amounts of AMPA and glyoxylate measured in plants from the resistant and susceptible L. rigidum populations (Table 2). Therefore, AMPA depletion and glyoxylate accumulation were only observed in susceptible L. rigidum, but not in resistant L. rigidum. However, the amounts of non-metabolized glyphosate were similar across populations, these results suggesting that glyphosate metabolism is not likely involved in the resistance (Table 2).
Glyphosate metabolism is not a frequent mechanism of resistance among weeds (Duke 2011). At this date, only Digitaria insularis (L.) Fedde and C. canadensis have been described as species having evolved resistance and able to transform glyphosate to non-toxic compounds (De Carvalho et al. 2012). Glyphosate metabolism is important in some naturally tolerant weeds and has been demonstrated in Ipomoea purpurea (L.) Roth, Convolvulus arvensis L., Clitoria ternatea L., Neonotonia wightii (Wight & Arn.) Verdc., and Mucuna pruriens (L.) DC. var. utilis (Wall. ex Wight) Baker ex Burck. All those species can degrade the herbicide glyphosate and produce high amounts of AMPA, glyoxylate, sarcosine, and glycine (Duke 2011; Cruz-Hipolito et al. 2011; Rojano-Delgado et al. 2012). However, many different factors may have influenced the results of metabolic events after glyphosate application to those different weeds, including analysis time after treatment, rates of metabolite formation and degradation, and possible endophyte contribution to metabolism. Anyway, our results suggest that glyphosate metabolism is not involved or plays a minor role in the resistance of the L. rigidum biotype we studied because glyoxylate accumulation in the susceptible L. rigidum population did not match a corresponding glyphosate metabolism. The same was reported in C. canadensis from USA (Dinelli et al. 2006).
3.5 EPSPS gene sequencing
To determine whether mutation in the EPSPS gene could explain the resistance to glyphosate in the studied L. rigidum biotype, the gene from both susceptible and resistant L. rigidum biotypes was partially sequenced. A fragment of 543 bp cDNA was amplified and compared with databases using the BLASTn and BLASTp algorithms. Results showed a high homology with the EPSPS gene from L. rigidum (GenBank: AAK20397.1) and Eleusine indica (GenBank: AJ417033.1). Protein alignment of the predicted EPSPS fragments from S and R biotypes of L. rigidum showed 88.4 and 89.1 % of protein similarity, respectively, to that of E. indica. Comparison of the EPSPS gene fragment between resistant and susceptible biotypes showed a mutation in the resistant biotype at position 106 in the protein amino acid sequence (Fig. 3). This mutation consisted in the presence of the AGC (serine) codon instead of CCG (proline) in the S biotype.
Amino acid substitutions of proline to serine, proline to threonine, proline to alanine, and proline to leucine at position 106 of the EPSPS protein were found to confer glyphosate resistance in grass weeds (Perez-Jones et al. 2007; Wakelin and Preston 2006; Baerson et al. 2002). Hence, in the EPSPS protein reported here, the same proline to serine amino acid substitution as described by Perez-Jones et al. 2007 is likely to take a part in glyphosate resistance in the resistant L. rigidum population.
Unlike glyphosate-resistant crops, where a prokaryotic enzyme-efficient, glyphosate-resistant EPSPS gene has been introduced, EPSPS mutations resulting in glyphosate-resistant weed biotypes reduce the affinity for substrates and the enzymatic efficiency (Preston et al. 2009). Thus, proline-to-serine and proline-to-threonine mutations not only confer a moderate glyphosate resistance in weed biotypes but additionally reduce the affinity of EPSPS for PEP binding (Shaner et al. 2011), decreasing fitness. This low efficiency in conferring resistance may explain the unusual pool of mechanisms of glyphosate resistance observed in the resistant populations. This accumulation of mechanisms of resistance has been already described in other glyphosate-resistant L. rigidum biotypes (Preston et al. 2009).
4 Conclusion
The above results indicate that resistance to glyphosate in the resistant population of L. rigidum is partly due to reduced absorption and translocation of glyphosate relative to the susceptible population, as well as to a mutation in the gene coding for the EPSPS.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. doi:10.1016/S0022-2836(05)80360-2
Baerson SR, Rodriguez DJ, Tran M, Feng Y, Biest NA, Dill GM (2002) Glyphosate-resistant goosegrass. Identification of a mutation in the target enzyme 5-enolpyruvylshikimate-3-phosphate synthase. Plant Physiol 129:1265–1275. doi:10.1104/pp. 001560
Collavo A, Sattin M (2014) First glyphosate-resistant Lolium spp. biotypes found in a European annual arable cropping system also affected by ACCase and ALS resistance. Weed Res 54:325–334. doi:10.1111/wre.12082
Cruz-Hipolito H, Rojano-Delgado A, Domínguez-Valenzuela JA, Heredia A, De Castro MDL (2011) Glyphosate tolerance by Clitoria ternatea and Neonotonia wightii plants involves differential absorption and translocation of the herbicide. Plant Soil 347:221–230. doi:10.1007/s11104-011-0840-9
De Carvalho LB, Alves PLDCA, González-Torralva F, Cruz-Hipolito HE, Rojano-Delgado AM (2012) Pool of resistance mechanisms to glyphosate in Digitaria insularis. J Agric Food Chem 60:615–622. doi:10.1021/jf204089d
Dinelli G, Marotti I, Bonetti A, Minelli M, Catizone P (2006) Physiological and molecular insight on the mechanisms of resistance to glyphosate in Conyza canadensis (L.) Cronq. biotypes. Pestic Biochem Physiol 86:30–41. doi:10.1016/j.pestbp.2006.01.004
Duke SO (2011) Glyphosate degradation in glyphosate-resistant and -susceptible crops and weeds. J Agric Food Chem 59:5835–5841. doi:10.1021/jf102704x
Favier T, Gauvrit C (2007) Premier cas de résistance au glyphosate en France. AFPP-Vingtième conférence du Columa, Dijon, pp 233–237
Franz JE, Mao MK, Sikorski JA (1997) Glyphosate: a unique global herbicide. ACS American Chemical Society, Washington, DC, 653 p
Gaines TA, Zhang W, Wang D, Bukun B, Chisholm ST (2010) Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc Natl Acad Sci U S A 107:1029–1034. doi:10.1073/pnas.0906649107
Ge X, André d'Avignon D, Ackerman JJH, Sammons RD (2010) Rapid vacuolar sequestration: the horseweed glyphosate resistance mechanism. Pest Manag Sci 66:345–348. doi:10.1002/ps.1911
Geiger DR, Fuchs MA (2002) Inhibitors of aromatic amino acid biosynthesis (glyphosate). In: Böger P, Wakabayashi K, Hirai K (eds) Herbicide classes in development: mode of action, targets, genetic engineering, chemistry. Springer, Berlin, pp 59–85, ISBN 978-3-642-59416-8
González-Torralva F, Cruz-Hipolito H, Bastida F, Mülleder N, Smeda RJ (2010) Differential susceptibility to glyphosate among the Conyza weed species in Spain. J Agric Food Chem 58:4361–4366. doi:10.1021/jf904227p
González-Torralva F, Gil-Humanes J, Barro F, Brants I, De Prado R (2012) Target site mutation and reduced translocation are present in a glyphosate-resistant Lolium multiflorum Lam. biotype from Spain. Plant Physiol Biochem 58:16–22. doi:10.1016/j.plaphy.2012.06.001
Heap I (2015) International survey of herbicide resistant weeds. Available: http://www.weedscience.org. Accessed Jan 2015
Michitte P, De Prado R, Espinoza N, Ruiz-Santaella JP, Gauvrit C (2007) Mechanisms of resistance to glyphosate in a ryegrass (Lolium multiflorum) biotype from Chile. Weed Sci 55:435–440. doi:10.1614/WS-06-167.1
Nandula VK, Reddy KN, Poston DH, Rimando AM, Duke SO (2008) Glyphosate tolerance mechanism in Italian ryegrass (Lolium multiflorum) from Mississippi. Weed Sci 56:344–349. doi:10.1614/WS-07-115.1
Owen MDK (2001) World maize/soybean and herbicide resistance. In: Powles SB, Shaner DL (eds) Herbicide resistance and world grains. CRC Press, Boca Raton, pp 101–163, ISBN: 0-8493229-7
Perez-Jones A, Park KW, Polge N, Colquhoun J, Mallory-Smith CA (2007) Investigating the mechanisms of glyphosate resistance in Lolium multiflorum. Planta 226:395–404. doi:10.1007/s00425-007-0490-6
Powles SB, Yu Q (2010) Evolution in action: plants resistant to herbicides. Annu Rev Plant Biol 61:317–347. doi:10.1146/annurev-arplant-042809-112119
Preston C, Wakelin AM, Dolman FC, Bostamam Y, Boutsalis P (2009) A decade of glyphosate-resistant Lolium around the world: Mechanisms, genes, fitness, and agronomic management. Weed Sci 57:435–441. doi:10.1614/WS-08-181.1
Rojano-Delgado AM, Ruiz-Jiménez J, De Castro MDL, De Prado R (2010) Determination of glyphosate and its metabolites in plant material by reversed-polarity CE with indirect absorptiometric detection. Electrophoresis 31:1423–1430. doi:10.1002/elps.200900583
Rojano-Delgado AM, Cruz-Hipolito H, De Prado R, Luque De Castro MD, Franco AR (2012) Limited uptake, translocation and enhanced metabolic degradation contribute to glyphosate tolerance in Mucuna pruriens var. utilis plants. Phytochemistry 73:34–41. doi:10.1016/j.phytochem.2011.09.007
Shaner DL (2009) Role of translocation as a mechanism of resistance to glyphosate. Weed Sci 57:118–123. doi:10.1614/WS-08-050.1
Shaner DL, Lindenmeyer RB, Ostlie MH (2011) What have the mechanisms of resistance to glyphosate taught us? Pest Manag Sci 68:3–9. doi:10.1002/ps.2261
Singh BK, Shaner DL (1998) Rapid determination of glyphosate injury to plants and identification of glyphosate-resistant plants. Weed Technol 12:527–530
Wakelin AM, Preston C (2006) A target-site mutation is present in a glyphosate-resistant Lolium rigidum population. Weed Res 46:432–440. doi:10.1111/j.1365-3180.2006.00527.x
Yu Q, Abdallah I, Han H, Owen M, Powles S (2009) Distinct non-target site mechanisms endow resistance to glyphosate, ACCase and ALS-inhibiting herbicides in multiple herbicide-resistant Lolium rigidum. Planta 230:713–723. doi:10.1007/s00425-009-0981-8
Acknowledgments
The authors thank Thierry Favier, who discovered this resistance case, for providing the L. rigidum seeds. This research was partly funded by the Agrochemicals and Environment Association.
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Fernandez, P., Gauvrit, C., Barro, F. et al. First case of glyphosate resistance in France. Agron. Sustain. Dev. 35, 1469–1476 (2015). https://doi.org/10.1007/s13593-015-0322-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13593-015-0322-1