Abstract
The novel Coronavirus, COVID-19, pandemic is being considered the most crucial health calamity of the century. Many organizations have come together during this crisis and created various Deep Learning models for the effective diagnosis of COVID-19 from chest radiography images. For example, The University of Waterloo, along with Darwin AI—a start-up spin-off of this department, has designed the Deep Learning model ‘COVID-Net’ and created a dataset called ‘COVIDx’ consisting of 13,975 images across 13,870 patient cases. In this study, COGNEX’s Deep Learning Software, VisionPro Deep Learning™, is used to classify these Chest X-rays from the COVIDx dataset. The results are compared with the results of COVID-Net and various other state-of-the-art Deep Learning models from the open-source community. Deep Learning tools are often referred to as black boxes because humans cannot interpret how or why a model is classifying an image into a particular class. This problem is addressed by testing VisionPro Deep Learning with two settings, first, by selecting the entire image as the Region of Interest (ROI), and second, by segmenting the lungs in the first step, and then doing the classification step on the segmented lungs only, instead of using the entire image. VisionPro Deep Learning results: on the entire image as the ROI it achieves an overall F score of 94.0%, and on the segmented lungs, it gets an F score of 95.3%, which is better than COVID-Net and other state-of-the-art open-source Deep Learning models.
Similar content being viewed by others
Introduction
The novel coronavirus disease, named COVID-19 by the World Health Organization—is caused by a new coronavirus class known as SARS-CoV2 (Severe Acute Respiratory Syndrome Coronavirus 2). It is a single-stranded RNA (ribonucleic acid) virus that causes severe respiratory infections. The first COVID-19 cases were reported in December 2019, in Wuhan, Hubei province, China [1]. As the virus has since spread worldwide, it has been given the status of pandemic by the World Health Organization. As of 16th February,2021, 12:00 GMT, 110 million people have been infected, and 2.4 million people have died due to COVID-19 [2]. One of the best solutions has been detecting the virus in its early stages and then isolating the infected individuals by quarantining them, thus preventing healthy people from becoming infected.
In many cases, real-time reverse transcriptase-polymerase chain reaction (RRT-PCR) of nasopharyngeal swabs has been used for diagnosis [3]. The RT-PCR throat swabs are collected from patients with COVID-19, and the RNA is then extracted. This process takes over two hours to complete and has a long turnaround time with limited sensitivity. The best alternative is to detect images of COVID-19 from radiology scans [4,5,5] (chest X-ray images and chest computed tomography (CT) images). The advantages of using chest X-rays over CT images are as follows: X-ray imaging systems are much more widely available than CT imaging systems, they are cost-effective, and digital X-ray images can be analysed at the point of acquisition, thus making the diagnosis process extremely quick [6].
X-ray images are grayscale. In medical imaging terms, these are images with values ranging from 0 to 255, where 0 corresponds to the completely dark pixels, and 255 corresponds to the completely white pixels. Different values on the X-ray image correlate to different areas of density. The different values are—dark: locations in the body which are filled with air are going to appear black, dark grey: subcutaneous tissues or fat, light grey: soft tissues like the heart and blood vessels, off white: bones such as the ribs, bright white: presence of metallic objects such as pacemakers or defibrillators. The way that physicians interpret an image is by looking at the borders between the different densities. The ribs appear off-white because they are dense tissues, but since the lungs are filled with air, the lungs appear dark. Similarly, below the lung is the hemidiaphragm, which is a soft tissue and hence appears light grey. This helps to find the location and extent of the lungs. If two objects with different densities are close to each other, they can be demarcated in an X-ray image. If something happens in the lungs, such as pneumonia, the air-dense lungs change into water-dense lungs. This causes the demarcation lines to fade since the pixel densities start closing in on the grayscale bar [7].
About 20% of patients infected with COVID-19 develop pulmonary infiltrates and some develop very serious abnormalities [8]. The virus reaches the lungs’ gas exchange units and infects alveolar type 2 cells [9, 10]. The most frequent CT abnormalities observed are ground-glass opacity, consolidation, and interlobular septal thickening in both lungs [11]. However, due to infection control issues related to patient transport to CT rooms, problems encountered with CT room decontamination, and the lack of CT scanner availability in different parts of the world, portable chest X-rays are likely to be one of the most common modalities for the identification and follow-up of COVID-19 lung abnormalities [12]. Hence, a significant number of expert radiologists who can interpret these images are needed. Due to the ever-increasing number of cases of COVID-19 infection, it is becoming more difficult for radiologists to keep up with this demand. In this scenario, Deep Learning techniques have proven to be beneficial in both classifying abnormalities from lung X-ray images and aiding the radiologists to accurately predict COVID-19 cases in a reduced time frame.
While many studies have demonstrated success in detecting images of COVID-19 using Deep Learning with both CT scans and X-rays, most of the Deep Learning architectures require extensive programming. Moreover, most of the architectures fail to showcase whether the Deep Learning model is being triggered by abnormalities in the lungs or artefacts not related to COVID-19. Due to the absence of a GUI (Graphical User Interface) with most of these Deep Learning models, it is difficult for radiologists, who lack knowledge in Deep Learning or programming, to use these models, let alone train them. Therefore, we showcase an already existing Deep Learning software with a very intuitive GUI, which can be used as a pre-trained software or can even be trained on new data from particular hospitals or research centres.
COGNEX VisionPro Deep Learning™ is a Deep Learning vision software, from COGNEX Corporation (Headquarters: Natick, MA, United States). It is a field-tested, optimised, and reliable software solution based on a state-of-the-art set of machine learning algorithms. VisionPro Deep Learning combines a comprehensive machine vision tool library with advanced Deep Learning tools.
In this study, we used the latest version—VisionPro Deep Learning 1.0—to aid in the classification of images as normal, non-COVID-19 (pneumonia), or COVID-19 chest X-rays. The results are compared with various state-of-the-art open-source neural networks. The VisionPro Deep Learning GUI, called COGNEX Deep Learning Studio, has three tools for image classification, segmentation, and location. It contains various Deep Learning architectures built within the GUI, to carry out specific tasks:
-
1.
Green Tool—This is the Classify tool. It is used to classify objects or complete scenes. It can be used to classify defects, cell types, images of different labels, or different types of test tubes used in laboratories. The Green tool learns from the collection of labelled images of different classes and can then be used to classify images that it has not previously seen. This tool is similar to classification neural networks such as VGG [13], ResNet [14] and DenseNet [15].
-
2.
Red Tool—This is the Analyse tool. It is used for segmentation and defect/anomaly detection; for example, to aid in the detection of anomalies in blood samples (clots), incomplete or improper centrifugation, or sample quality management. The Red tool is also used to segment specific regions, such as defects or areas of interest. The Red tool comes with the option of using either Supervised Learning or Unsupervised Learning for segmentation and detection. This is similar to the segmentation neural network, such as U-Net [16].
-
3.
Blue Tool—(a) This is the Feature Localisation and Identification tool. The Blue tool finds complex features and objects by learning from labelled images. It has self-learning algorithms that can locate, classify, and count the objects in an image. It can be used for locating organs in X-ray images or cells on a microscopic slide. (b) The Blue tool also has a Read feature. It is a pre-trained model that helps to decipher severely deformed and poorly etched words and codes using optical character recognition (OCR). This is the only pre-trained tool. All other tools need to be trained on images first to get results.
For the classification of COVID-19 images, two settings are used:
-
1.
Green tool for classification of the entire chest X-ray images
-
2.
Red tool for segmentation of the lungs, and then a subsequent Green tool classifier just run on the segmented lungs to make sure the Deep Learning software predicts its results based on the lungs only.
Method
Dataset
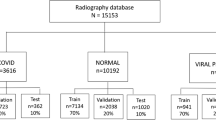
The open access benchmark dataset called COVIDx was used for training the various models [17]. The dataset contains a total of 13,975 Chest X-ray images from 13,870 patients. The dataset is a combination of five different publicly available datasets. According to the authors [17] of COVID-Net, COVIDx is one of the largest open-source benchmark datasets in terms of the number of COVID-19 positive patient cases.
These five datasets were used by the authors of COVID-Net to generate the final COVIDx dataset:
-
(a)
Non-COVID 19 pneumonia patient cases and COVID-19 cases from the COVID-19 Image Data Collection [18],
-
(b)
COVID-19 patient cases from the Fig. 1 COVID-19 Chest X-ray Dataset [19],
-
(c)
COVID-19 patient cases from the ActualMed COVID-19 Chest X-ray Dataset [20],
-
(d)
Patient cases who have no pneumonia (that is, normal) and non-COVID-19 pneumonia patient cases from RSNA Pneumonia Detection Challenge dataset [21],
-
(e)
COVID-19 patient cases from the COVID-19 radiography dataset [22].
Chest X-ray image distribution of each class: normal (no infection), non-COVID-19 (Pneumonia), and COVID-19 images. In the training set, there are 7966 images belonging to the ‘Normal’ class, 5451 images in the ‘Non-COVID-19’ class, and 258 ‘COVID-19’ images. In the test set, there is an equal distribution of 100 images across all the three classes. Horizontal axis represents the different categories or classes, and the vertical axis represents the number of images in each of these categories
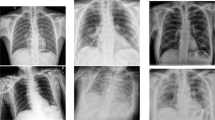
The idea behind using these five datasets was that these are all open-source COVID-19/Pneumonia Chest X-ray datasets, so they can be accessed by everyone in the research community and by the general public, and also add variety to the dataset. However, the lack of COVID-19 Chest X-ray images made the dataset highly imbalanced. Of the 13,975 images, the data were split into 13,675 training images and just 300 test images. The data were divided across three classes: (1) normal (for X-rays which did not contain Pneumonia or COVID-19), (2) non-COVID-19/Pneumonia (for X-rays, which had some form of bacterial or viral pneumonia, but not COVID-19), and (3) COVID-19 (for X-rays which were COVID-19 positive). In the training set, there were 13,675 images, with 7966 of those belonging to the Normal class, 5451 images belonging to the Non-COVID-19/Pneumonia class, and only 258 images in the COVID-19 class. The test set was a balanced set, with each of the three classes having 100 of each image type [17]. Fig. 2 shows three X-ray images from each class in the dataset.
The authors of COVID-Net have shared the dataset generating scripts, for the construction of the COVIDx dataset for public access available at the following link—https://github.com/lindawangg/COVID-Net [17]. The python notebook ‘create_COVIDx_v3. ipynb’ was used to generate the dataset. The text files ‘train_COVIDx3.txt’ and ‘test_COVIDx3.txt’ contains the file names used in the training and test set, respectively. It was then tested with VisionPro Deep Learning, and the results were compared with COVID-Net results and other open-source Convolutional Neural Network (CNN) architectures such as VGG [13] and DenseNet [15]. Tensorflow [33] (developed by Google Brain Team [34]) library was used to generate and train the open-source CNN architectures
Examples of the chest X-ray images belonging to the different classes. The class numbers are shown along the vertical axis. Class 1: normal images, Class 2: non-COVID-19 (Pneumonia) images and Class 3: COVID-19 images. All images belong to the training set of the COVIDx dataset [17]
Pre-processing Data
The scripts for generating the COVIDx dataset were used to merge the five datasets together and separate the images into training and test folders. Along with the images, the script also generated two text files containing the names of images belonging to the training and test folders, and their class labels [17].
To simplify classification, a python script was used to convert the ‘.txt’ files into ‘pandas’ data frames and then finally converted to ‘.csv’ files for better understanding. Next, another python script was created to rename all of the X-ray images of the training and test folders according to their class labels and store them in new training and test directories. Since the goal was classification of the X-ray images, renaming the images made it easier to interpret the images directly from their file names, rather than consulting a ‘.csv’ file every time. Finally, we have all the 13,975 images in train and test folders, with their file names containing the class labels.
-
(a)
COGNEX VisionPro Deep Learning
Unlike most other Deep Learning architectures VisionPro Deep Learning does not require any pre-processing of the images. The images can be fed directly into the GUI, and the software automatically does the pre-processing, before starting to train the model.
Since the COVIDx dataset is a combination of various datasets, the images have different colour depths, and VisionPro Deep Learning GUI found 326 anomalous images. Training could have been done by keeping the anomalous images in the dataset, but it might have reduced the overall F score of the model. Therefore, we normalised the colour depth of all COVIDx images to 24-bit colour depth using external software, IrfanView (open source: irfanview.com). Then, the images were added into the VisionPro Deep Learning GUI.
No other pre-processing steps are necessary with VisionPro Deep learning, such as image augmentation or setting class weights or oversampling of the imbalanced classes, which are necessary for the training the other open-source CNN models. Once the images are fed into the VisionPro Deep learning GUI, they are ready to be trained.
-
(b)
Open Source Convolutional Neural Network (CNN) Models
Before training the CNN models, such as VGG [13] or DenseNet [15], it was necessary to execute some pre-processing steps, such as resizing, artificial oversampling of the classes with fewer images, image standardisation and finally data augmentation. First, the images were resized to 256 × 256 pixels. The entire training was done on an Nvidia 2080 GPU, as this was found to be the perfect image size to not run into ‘GPU memory errors. Once the images were resized, and the images and labels loaded together, it was necessary to oversample images which belong to the classes having fewer images, that is, for the Non-COVID-19 and COVID-19 classes. For oversampling, random artificial augmentations were carried out, such as rotation (− 20° to + 20°), translation, horizontal flip, Gaussian blur, and adding external noise. All of these were applied randomly using the ‘random’ library in python. Then, all of the X-ray images were standardised to have values with a mean of zero and a standard deviation of 1. This was done, keeping in mind that standardisation helps the Deep Learning network to learn much faster. Finally, data augmentation was added to all classes, irrespective of the number of images belonging to those classes. Augmentations include rescaling, height and width shifting, rotating, shearing and zooming. After all of these pre-processing steps, the images were ready to be fed into the deep neural networks.
Classification using VisionPro Deep Learning
The goal of the study was the classification of normal, non-COVID-19(Pneumonia) and COVID-19 X-ray images. For classification, VisionPro Deep Learning uses the Green tool. Once the images were loaded and labelled, they were ready for training. In VisionPro Deep Learning, the Region of Interest (ROI) of the images can be selected. Thus, it is possible to reduce the edges by 10–20% to remove artefacts like the letters or borders, which are usually at the edges of the images. In this case, the entire images were used without cropping the edges because many images have the lungs towards the edge, and we did not want to remove essential information.
To feed the images into VisionPro Deep learning, the images did not need to be resized. Images of all resolutions and aspect ratios can be fed into the GUI, and the GUI does the pre-processing automatically before starting the training. In VisionPro Deep Learning, the Green tool has two subcategories, High-detail and Focussed. Under High-detail there are several options such as sizes of model architectures—small, normal, large and extra-large models, which can be selected for training the model. We train the network using the High-detail subcategory and selecting the ‘Normal’ size model.
Out of the 13,675 images, 80% of the images are used for training. The VisionPro Deep Learning suite automatically selects the other 20% images for validation. Both the training and validation sets are randomly selected by the VisionPro Deep Learning suite. The user just needs to specify the train-validation split. The maximum number of epoch counts was selected to be 100. There are options of selecting the minimum epochs and patience for which the model will train, but this was not selected. Once these are selected, training is started by clicking on the ‘brain’ icon on the green tool, as seen in Fig. 3.
The VisionPro Deep Learning GUI loaded with the X-ray images from the COVIDx dataset [17]. On the left of the GUI, there are options to select various parameters for training the model, such as model type, model size, epoch count, minimum epochs and patience, train and validation split, class weights, threshold, heatmap and the different data augmentation options of flip, rotation, contrast, zoom, brightness, sharpen, blur, distortion and noise. In the middle, the selected image is shown. On the right, thumbnails of all the images in the training and test set are shown. On the top, there is the tool selection option. In the figure, the green tool has been selected for classification. Clicking on the ‘brain’ shaped icon in the green tool, starts the training of the model
Segmentation and improved classification using VisionPro Deep Learning
The Green tool is used to classify entire X-ray images, but for the identification of images of COVID-19, the Deep Learning model needs to focus on the lungs, and not the peripheral bones, organs and soft tissues. The model must make its predictions exclusively based on the lungs and not on the differences in spinous process, clavicles, soft tissues, ornaments worn around the patient’s neck or even the background. This way we can be sure that the model is classifying based entirely on the normal and infected lungs. Therefore, segmentation of the lungs from each image makes sure that the model trains only on these segmented lungs, and not on the entire image. To implement this, the VisionPro Deep Learning Red tool is used. The Red tool is used to segment the images, such that only the lungs are visible to the Deep Learning model for training. To achieve this, 100 images of the training set are manually masked using the ‘Region selection’ option in the Red Tool. The training set consists of 13,675 images, but a manual masking of 100 images is enough to train the model. Once the manual masking is done on the 100 images, the Red tool is trained. After training is completed, the VisionPro Deep Learning GUI has all the training and test images properly masked, as seen in Figs. 4 and 5, such that only the lungs are visible. Anything outside the lungs is treated as outside the ROI and is not used in classification. The Red tool is added in the same environment as the previous green tool and there is no need to create a new instance for segmentation.
Once all of the images are segmented, a Green classification tool is implemented after the Red tool. The Green tool is then used to start the classification (similar to Step 3 of “Method”), but this time exclusively on the segmented lungs and not on the entire images.
Classification using VGG network
The VGG [13] network is a deep neural network and is still one of the state-of-the-art Deep Learning models used in image classification.
We used the 19-layer VGG 19 model for training using transfer learning on the COVIDx dataset. VGG takes an image of size 224 × 224 pixels. Pre-processing of the images was performed automatically by calling ‘preprocess input’ from the VGG19 model in TensorFlow. The ‘preprocess input’ is fed into the ‘ImageDataGenerator’ from TensorFlow (Keras). ‘ImageNet’ weights are used for training. The COVIDx dataset was also resampled as stated in 2 (b) of the “Method”. This ensures that all classes have similar number of images, to avoid the model favouring a particular class during training. The VGG19 architecture uses 3 × 3 convolutional filters which performs much better than the older AlexNet [23] models. All of the activation functions used in the hidden layers are ReLU (Rectified Linear Units) [24]. After the VGG architecture, we add four fully connected layers with 1024 nodes each. All four layers use the ReLU activation function and L2 regularization [25, 27]. To provide better regularisation, after each of these layers a Dropout is set. The final layer is a fully connected layer of three nodes for the classification of the 3 classes. The final layer has the activation function ‘SoftMax’ [27].
In the pre-processing steps, the labels of the images are not ‘one-hot encoded’ but kept as three distinct digits. So instead of using ‘categorical cross entropy’ [28], which is commonly used when the labels are one-hot encoded, the ‘sparse categorical cross entropy’ is used as the loss function. ‘Adam’ [29] optimiser is used with learning rate scheduling, such that the learning rate decreases after every thirty epochs. During training, several call-backs are set, such as saving the model each time the validation loss decreases, and using early stopping, to stop the training when the validation loss does not improve even after several epochs. The epoch count is set to 100. For training, batches of 32 images are fed to the model at once. Once all of these hyperparameters are set, training of the model is started. After training completion, the programme is set to plot the confusion matrix and give results on the various evaluation metrics, based on which the various models are compared.
Classification using ResNet
One of the bottlenecks of the VGG network is that it does not go too deep as it starts losing generalisation capability the deeper it goes. To overcome this problem ResNet or Residual Network [14] is chosen. The ResNet architecture consists of several residual blocks with each block having several convolutional operations. The implementation of skip connections makes the ResNet better than VGG. The skip connections between layers add the outputs from previous layers to the outputs of the stacked layers. This allows the training of deeper networks. One of the problems that ResNet solves is the vanishing gradient problem [30].
For training the COVIDx dataset, we use the 50-layer ResNet50V2 (Version 2) architecture. We use transfer learning to train the model, and then add eight fully connected layers with L2 regularisation followed by Dropouts for better regularisation. All of the other settings and hyperparameters are kept similar to the training of the VGG19 network (“Method”, part 5).
Classification using DenseNet
DenseNet (Dense Convolutional Network) [15] is an architecture which focuses on making the Deep Learning networks go even deeper, while at the same time making them more efficient to train, using shorter connections between the layers. DenseNet is a convolutional neural network where each layer is connected to all other layers that are deeper in the network, that is, the first layer is connected to the 2nd, 3rd, 4th and so on, the second layer is connected to the 3rd, 4th, 5th and so on. Unlike ResNet [14], it does not combine features through summation but combines the features by concatenating them. So, the ‘ith’ layer has ‘i’ inputs and consists of feature maps of all its preceding convolutional blocks. It therefore requires fewer parameters than traditional convolutional neural networks.
To train the COVIDx dataset, we use the 121 layered DenseNet121 architecture. We use transfer learning to train the model, and then add eight fully connected layers with L2 regularisation followed by Dropouts for better regularisation. All the other settings and hyperparameters are kept similar to the training of the VGG19 network (“Method”, part 5).
Classification using Inception Network
Inception Network [31] has been developed with the idea of going even deeper with convolutional blocks. Very deep networks are prone to overfitting, and it is hard to pass gradient updates throughout the entire network. Also, images may have huge variations, so choosing the right kernel size for convolution layers is hard. To address these problems, Inception network is one of the best possible networks. Inception network version 1 has multiple sizes of filters in the same level. It has various connections of three different sizes of filters of 1 × 1, 3 × 3, 5 × 5, with max pooling in a single inception module. All of the outputs are concatenated and then sent to the next inception module
The segmented lungs after training the Red tool in VisionPro Deep Learning. Anything outside the segmented lungs is not considered to be part of the Region of Interest (ROI) and is not used for classification. This makes sure that VisionPro Deep Learning trains only on the lungs and not on the artefacts around it
To train the COVIDx dataset, we use the 48 layered InceptionV3 [32] architecture, which also includes 7 × 7 convolutions, Batch Normalisation and Label smoothing in addition to the Inception version 1 modules. We use transfer learning to train the model, and then add eight fully connected layers with L2 regularization followed by Dropouts for better regularization. All of the other settings and hyperparameters are kept similar to the training of the VGG19 network (“Method”, part 5).
Results
Evaluation Metrics
In medical imaging, since the decisions are of high impact, it is very important to understand exactly which evaluation metrics are necessary to decide whether a model works on a patient or not. Accuracy of a model is not the best metric for deciding whether the model is fit for a patient. Rather, it is important to look into other evaluation metrics such as sensitivity, predictive values and overall F-scores. First, the confusion matrix is plotted for the 300 test images, for all of the models that we use for the comparison.
Figure 6 shows the confusion matrix of VisionPro Deep Learning on the entire ROI. VisionPro Deep learning GUI does not display numbers of correctly classified or misclassified images on the confusion matrix, but if any point on the confusion matrix is clicked, it displays not only the number of images in that category, but also all the images belonging to that category, with the prediction percentage and whether the prediction it made is correct or not. Below the confusion matrix, it displays all of the evaluation metrics of recall (sensitivity), precision (positive predictive value) and F-scores. This table contains the number of labelled images, which shows the number of images in each class in the test set. The ‘Found’ column shows the number of images that VisionPro Deep Learning thinks should belong in those classes. A report can also be generated on all of the test images, as seen in Fig. 7 and 8. It shows a small snippet of six COVID-19 positive images from the test set. The report contains details of the 300 test images, including the filename, the image, the original label as ‘Labelled’, and the predictions made by VisionPro Deep learning as ‘Marked’, with the percentage of confidence of prediction on each class. If the prediction is different from that of the label, then it is marked in red.
Left: confusion matrix on the 300 test images COGNEX VisionPro Deep Learning with entire ROI selected. Confusion matrix on the 300 test images. COGNEX VisionPro Deep Learning with entire ROI selected. Right: interpretation of the confusion matrix. VisionPro Deep learning GUI does not display numbers of correctly classified or misclassified images on the confusion matrix, but if any point on the confusion matrix is clicked, it displays not only the number of images in that category, but also all of the images belonging to that category, with the prediction percentage and whether the prediction it made is correct or not
A snippet of the report generated on the 300 test images by VisionPro Deep Learning with the entire image selected as the ROI. The report contains the confusion matrix with the evaluation metrics: sensitivity (recall), positive predictive value (precision) and F score for each class. The test images are also shown with the correct labels, the predicted labels and the confidence percentage of each class. In this image, 5 images are classified correctly, and 1 image misclassified (marked in red)
Misclassification results Of the 300 test images, VisionPro Deep Learning classified 18 images incorrectly with the entire ROI selected, and 16 images incorrectly with the segmented lungs. COVID-Net had classified 20 images incorrectly [17]. VGG19 [13], ResNet50V2 [14], Densenet121 [15], and InceptionV3 [32] networks made 47, 37, 41, 26 misclassifications, respectively. VisionPro Deep Learning has fewer misclassifications than all of the open-source models in both the settings. Compared to COVID-Net [17], the performance of VisionPro Deep learning is similar with the entire image as the ROI and much better when using the segmented lungs .
Heatmaps are a great way to visualise predictions of the Deep Learning algorithm. They highlight exactly which parts of the image trigger the model to generate its predictions. Figure 9 shows the heatmaps generated by VisionPro Deep Learning on six COVID-19 images.
Four COVID-19 X-ray images from the test set of the COVIDx dataset along with the predicted heatmaps generated by VisionPro Deep Learning. Heatmaps can be a great indicator for radiologists to identify whether the predictions made by the Deep learning algorithm is based on actual infection or some artefacts
Confidence interval A confidence interval is a range of values that we are fairly sure the true value always lies in. Since the number of images in the test set was so small, with only 100 images in each class, we saw high confidence intervals in most of the cases, both with the open-source models, and with VisionPro Deep Learning. The best possible way to reduce the confidence interval is to increase the number of images in the test set, by a range, which lies somewhere in the thousands and not in the hundreds. Since the number of COVID-19 images was very small, and we wanted to make a one-to-one comparison with the results of COVID-Net [17], we used the same number of images provided in the test set of the COVIDx dataset. We calculated a 95% confidence interval on the predicted sensitivity and the positive predicted values, to determine a possible range of values by which the actual results may vary on the given test data. The confidence interval of the accuracy rates is calculated using the formula:
where z is the significance level of the confidence interval (the number of standard deviation of the Gaussian distribution), accuracy is the estimated accuracy (in our cases sensitivity, positive predictive value, and F score), and N (100 for each class) denotes the number of samples for that class. Here, we used the 95% confidence interval, for which the corresponding value of z is 1.96 [34].
Sensitivity
Sensitivity or Recall measures the true positive rate. It is the proportion of the true positives detected by a model to the total number of positives. The better the sensitivity, the better the model is at correctly identifying the infection.
For normal and COVID-19 classes, VisionPro Deep Learning significantly outperforms all other models, as seen in Table 1. For the Non-COVID-19 class COVID-Net [17] has the best results. VisionPro Deep Learning has a really good sensitivity to COVID-19 images: 95% for the images with the entire ROI selected, and 97% for images with the lungs segmented. Also, both settings of VisionPro Deep Learning showed 98% sensitivity for images belonging to the normal class.
Positive Predictive Values
Positive predictive value (PPV) or Precision shows the percentage of how many predictions selected by the model are relevant.
As seen in Table 2, DenseNet121 [15] has the best PPV for Normal images, VisionPro Deep Learning has the best PPV for Non-COVID-19 images and COVID-Net [17] has the best PPV for COVID-19 images. Although it is not the best in comparison, VisionPro Deep Learning still has a high PPV value for COVID-19 images, with 96.9% for the images with the entire ROI selected, and 97.0% for the images with the lungs segmented.
Overall F-scores
F score takes into consideration both the Sensitivity and PPV of a model. It can be considered as an overall score of the performance of the model.
As seen in Table 3, of the open-source architectures, InceptionV3 [32] has the best F score for all the three classes. When compared to InceptionV3, COVID-Net has a higher F score in the Non-COVID-19 and COVID-19 classes but is slightly lower in the Normal class. VisionPro Deep Learning on the entire image as the ROI outperforms all of the open-source architectures and COVID-Net, except in the Non-COVID-19 class, where the results are very close, with COVID-Net [17] having an F score of 92.6% and VisionPro Deep Learning having an F score of 92.2%. The setting with segmented lungs, VisionPro Deep Learning outperforms all the open-source models, COVID-Net [17] and even itself with the entire ROI selected. On all classes, it has the highest F score. It gets an F score of 95.6% for normal images, 93.3% for non-COVID-19/Pneumonia images and an F score of 97.0% for the COVID-19 images. In this setting, VisionPro Deep Learning is only classifying based on the lungs, so there are no artefacts, and the results obtained are highly focussed. This helps to overcome the black-box idea of Deep Learning results.
VisionPro Deep Learning has the best F-scores on COVID-19 images for both of the settings. On the entire ROI it has an F score of 96.0% and on the segmented lungs it has an F-scores of 97.0%. Overall, for all three classes, VisionPro Deep Learning achieves an F score of 94.0% for the entire image as the ROI, and an F score of 95.3% for the segmented lungs. The similarity of the results in both the settings and the heatmaps, show that even without the lungs being segmented, VisionPro Deep Learning is still predicting its classes based on the actual abnormalities. Figures 10 and 11 show the confusion matrix of the various open-source models and COVID-Net [18], respectively.
Confusion matrix on the 300 test images for the open-source architectures a VGG19 [14], b Resnet50 V2 [15], c Densenet121 [16], d Inception V3 [33]. Inception V3 has the best results, with the lowest number of false predictions. ResNet50 V2 has the next best result, followed by DenseNet121 and VGG19, respectively
Confusion matrix on the 300 test images for COVID-Net. Image from the original COVID-Net paper [17]. COVID-Net results are better than all of the open-source models that we use for training
VisionPro Deep Learning tested on a previous version of the COVIDx dataset. This dataset has many more images for the test set, in the Normal and Non-COVID-19 classes, but only 91 images in the COVID-19 class. We see the confidence interval improve significantly in classes with a higher number of test images
As expected, when comparing the confidence intervals, none of the models perform well due to the significantly lower number of images in each class in the test set.
We also tested VisionPro Deep Learning on a previous version of the COVIDx dataset, which has a total of 15,374 images. They were split into the following number of images in the training set: 7965 images in the normal class, 5459 images in the non-COVID-19 class and 380 images in the COVID-19 class, and the following number of images in the test set of each class: 885 images in the Normal class, 594 images in the Non-COVID-19 class and 91 images in the COVID-19 class. As seen in Table 4, due to the significantly higher number of images in the Normal and Non-COVID-19 class, the confidence interval significantly improves from the previous values ranging from 3–5% to just 1.0–2.4%. Figure 12 shows the results on this dataset on Cognex VisionPro Deep Learning. These results clearly indicate that as the number of test images increased, the confidence interval improved significantly. Similarly, as this dataset had only 91 images in the COVID-19 class, the confidence interval was similar to the previous results.
Also, Table 4 indicates that even when the number of images in the test set is significantly increased, the performance of VisionPro Deep Learning does not decrease, but rather still produces sensitivity, PPV and F-scores above 90% in all of the classes. If Table 4 is compared with the previous results, it can be seen that the results are very consistent for VisionPro Deep Learning, even with a change in the number of images in the training and test sets. Also, the results for the sensitivity, PPV and F-scores are very similar with the entire image as the ROI and also for the segmented lungs, further indicating that the predictions are made based on the lungs and not on the surrounding artefacts.
Similar Literature Study of Using Deep Learning Algorithms for Identification of COVID-19
Various other studies have been undertaken for the detection of COVID-19 from radiological images. One such study implements the idea of using Active Learning (AL), which implements Incremental Learning (IL), allowing the algorithm to self-learn over time in the presence of experts [34]. The aim of the study was to create a model which iteratively learns and adapts to new data without forgetting what it has previously learnt. Another study showed how their network performed equally well for both X-ray and CT images [35]. The study designed its own deep learning architecture which was trained on 336 Chest X-ray and 336 CT scan images. It reached a sensitivity of 97% and a precision of 94% on the dataset. A truncated form of the Inception network [36] achieved an accuracy of 99.96% while classifying COVID-19 positive cases from combined pneumonia and healthy cases, and an accuracy of 99.92% when classifying COVID-19 cases from combined pneumonia, tuberculosis and healthy chest X-rays. CoroNet [37], which is based on the Xception [38] architecture, was trained on another X-ray dataset and compared with COVID-Net [17]. CoroNet [37] achieved an accuracy of 89.6% on the dataset, while COVID-net [17] achieved an accuracy of 83.5%. COVID_MTNet [39] is another architecture which classifies and segments both Chest X-rays and CT scan images and obtains an accuracy of 94.67% on Chest X-rays and 98.78% on Chest CT scan images. In some cases, generative adversarial networks (GANs) [40], such as CycleGAN [41], were used to augment the minority class of COVID-19 images [42]. Several networks have also been designed to forecast the growth and spread of COVID-19 [43]. In fact, several books have been published which showcase systems and methods to prevent the further spread of COVID-19 using artificial intelligence, computer vision and robotics [44, 45].
Conclusion
In this study, we used COGNEX’s Deep Learning Software VisionPro Deep Learning (version 1.0) and compared its performance with other state-of-the-art Deep Learning architectures. VisionPro Deep Learning has an intuitive GUI, making the software very easy to use. Building applications requires no coding skills in any programming language, and little to no pre-processing is required, also decreasing the development time. Imbalanced data are automatically balanced within the software. Once the images are loaded into VisionPro Deep Learning and the correct tool is selected, the Deep Learning training can start. After the completion of training, it outputs a confusion matrix, along with various important metrics, such as precision, recall and F score. Additionally, a report can be generated that identifies all misclassified images. This makes it particularly suitable for radiologists, hospitals, and research workers to harness the power of Deep Learning without advanced coding knowledge. Moreover, as the results from this study indicates, the Deep Learning algorithms in VisionPro Deep Learning are robust and comparable or even better than the various state-of-the-art algorithms available today. The problem of Deep Learning algorithms being a “black box” can be overcome using a pipeline of tools, stacked sequentially to first segment the lungs and then classify only on the segmented lungs. It is like combining a U-Net [16] and Inception [31] model together. This ensures that the algorithm does not focus on any artefacts when generating its classification results. A heatmap can be generated to showcase exactly where the model is focussing when making the predictions, and with both settings, using the entire image as the ROI and classification on the segmented lungs, VisionPro Deep Learning achieves the highest overall F-scores, surpassing the results of the various open-source architectures.
In the future, more testing will be performed to understand how changing the number of training images or using augmentations in the training set affects the performance of VisionPro Deep Learning compared to the other open-source models. The software also gave F-scores of 99% on identification of COVID-19 from CT images [46].
This software is by no means a stand-alone solution in the identification of images of COVID-19 from Chest X-rays, but can help radiologists and clinicians to achieve a faster and understandable diagnosis using the full potential of Deep Learning, without the prerequisite of having to code in any programming language.
Data Availability
COVIDx dataset, which is created by The University of Waterloo, along with Darwin AI, is used and is available open source. It is available on the following link—https://github.com/lindawangg/COVID-Net [17]. Details on the dataset is given below, under "Method" (1. Dataset) in this manuscript.
Code availability
COGNEX VisionPro Deep Learning software is provided by COGNEX Corporation. The results from VisionPro Deep Learning is tested using this software. COVID-Net results are used from the original COVID-Net paper [17]. The code is available on—https://github.com/lindawangg/COVID-Net. Other open-source architectures such as VGG19 [13], ResNet V2 [14], DenseNet121 [15], and Inception V3 [32], are trained using transfer learning with the TensorFlow 2.0 library. The codes are available on—https://github.com/as4401s/COVID-19-X_ray-image-classification.
Abbreviations
- COVID-19:
-
Coronavirus Disease 2019
- SARS-CoV2:
-
Severe Acute Respiratory Syndrome Coronavirus 2
- ROI:
-
Region of interest
- RRT-PCR:
-
Real-time reverse transcriptase-polymerase chain reaction
- CT:
-
Computed tomography
- GUI:
-
Graphical user interface
- RNA:
-
Ribonucleic acid
References
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. (2020) ;395:497–506. https://doi.org/10.1016/S0140-6736(20)30183-5.
Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y, Li Y, Wang X, Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 Novel Coronavirus-infected pneumonia in Wuhan, China. J Am Med Assoc. (2020). https://doi.org/10.1001/jama.2020.1585.
Ng M-Y, Lee EY, Yang J, Yang F, Li X, Wang H, Lui MM, Lo CS-Y, Leung B, Khong P-L, Hui CK-M, Yuen K, Kuo MD. Imaging profile of the COVID-19 infection: radiologic findings and literature review. RadiolCardiothorac Imaging. (2020). https://doi.org/10.1148/ryct.2020200034.
Guan W, Ni Z, Hu Y, Liang W, Ou C, He J, Liu L, Shan H, Lei C, Hui DSC, Du B, Li L, Zeng G, Yuen K-Y, Chen R, Tang C, Wang T, Chen P, Xiang J, Zhong N. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. (2020). https://doi.org/10.1056/nejmoa2002032.
Mangal M, Kalia S, Rajgopal H, Rangarajan K, Namboodiri V, Banerjee S, Arora S. COVIDAID: COVID-19 detection using chest X-ray. (2020). http://arxiv.org/abs/2004.09803v1
Novelline R. Squire’s fundamentals of radiology. 5th ed. Cambridge: Harvard University Press; (1997). (ISBN 0–674–83339–2).
Mason RJ. Pathogenesis of COVID-19 from a cell biology perspective. EurRespir J. (2020). https://doi.org/10.1183/13993003.00607-2020.
Mossel EC, Wang J, Jeffers S, et al. SARS-CoV replicates in primary human alveolar type II cell cultures but not in type I-like cells. Virology. (2008);372:127–35. https://doi.org/10.1016/j.virol.2007.09.045.
Weinheimer VK, Becher A, Tonnies M, et al. Influenza A virus’s target type II pneumocytes in the human lung. J Infect Dis. (2012);206:1685–94. https://doi.org/10.1093/infdis/jis455.
Wu J, Wu X, Zeng W, Guo D, Fang Z, Chen L, Huang H, Li C. Chest CT findings in patients with coronavirus disease 2019 and its relationship with clinical features. Invest Radiol. (2020). https://doi.org/10.1097/RLI.0000000000000670.
Jacobi A, Chung M, Bernheim A, Eber C. Portable chest X-ray in coronavirus disease-19 (COVID-19): a pictorial review. Clin Imaging. (2020). https://doi.org/10.1016/j.clinimag.2020.04.001.
Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. In: 3rd International conference on learning representations, (2015) ICLR 2015—conference track proceedings.
He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proceedings of the IEEE computer society conference on computer vision and pattern recognition. (2016). https://doi.org/10.1109/CVPR.2016.90
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. In: Proceedings—30th IEEE conference on computer vision and pattern recognition, CVPR 2017. (2017). https://doi.org/10.1109/CVPR.2017.243
Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In: Lecture notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics). (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Wang L, Lin ZQ, Wong A. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci Rep. (2020) ;10:19549. https://doi.org/10.1038/s41598-020-76550-z.
Cohen JP, Morrison P, Dao L. COVID-19 image data collection. (2020) In: arXiv.
Chung A. Figure 1 COVID-19 chest X-ray data initiative. (2020). https://github.com/agchung/Figure1-COVID-chestxray-dataset.
Chung A. Actualmed COVID-19 chest x-ray data initiative. (2020). https://github.com/agchung/Actualmed-COVID-chestxray-dataset.
R.S. of North America, RSNA pneumonia detection challenge. (2019). https://www.kaggle.com/c/rsna-pneumonia-detection-challenge/data.
Chowdhury MEH, Rahman T, Khandakar A, Mazhar R, Kadir MA, Mahbub ZB, Islam KR, Khan MS, Iqbal A, Emadi NA, Reaz MBI, Islam MT. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access. (2020). https://doi.org/10.1109/ACCESS.2020.3010287.
Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Commun ACM. (2017). https://doi.org/10.1145/3065386.
Dahl GE, Sainath TN, Hinton GE. Improving deep neural networks for LVCSR using rectified linear units and dropout. In: ICASSP, IEEE international conference on acoustics, speech and signal processing—proceedings. (2013). https://doi.org/10.1109/ICASSP.2013.6639346
Cortes C, Mohri M, Rostamizadeh A. L2 regularization for learning kernels. (2009) In: Proceedings of the 25th conference on uncertainty in artificial intelligence, UAI 2009
van Laarhoven T. L2 regularisation versus batch and weight normalisation. (2017). http://arxiv.org/abs/1706.05350.
Goodfellow I, Bengio Y, Courville A. Deep learning. MIT Press. (2016). http://www.deeplearningbook.org
Zhang Z, Sabuncu MR. Generalized cross entropy loss for training deep neural networks with noisy labels. (2018) In: Advances in neural information processing systems. Cambridge: MIT Press.
Kingma DP, Ba J. Adam: a method for stochastic optimisation. (2014). http://arxiv.org/abs/1412.6980.
Veit A, Wilber M, Belongie S. Residual networks behave like ensembles of relatively shallow networks. (2016) In: Advances in neural information processing systems. Cambridge: MIT Press.
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A. Going deeper with convolutions. In: Proceedings of the IEEE computer society conference on computer vision and pattern recognition. (2015). https://doi.org/10.1109/CVPR.2015.7298594
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE computer society conference on computer vision and pattern recognition. (2016). https://doi.org/10.1109/CVPR.2016.308
Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M, Kudlur M, Levenberg J, Monga R, Moore S, Murray DG, Steiner B, Tucker P, Vasudevan V, Warden P, Zheng X. TensorFlow: a system for large-scale machine learning. (2016) In: Proceedings of the 12th USENIX symposium on operating systems design and implementation, OSDI 2016
Santosh KC. AI-driven tools for coronavirus outbreak: need of active learning and cross-population train/test models on multitudinal/multimodal data. J Med Syst. (2020). https://doi.org/10.1007/s10916-020-01562-1.
Mukherjee H, Ghosh S, Dhar A, et al. Deep neural network to detect COVID-19: one architecture for both CT scans and chest X-rays. ApplIntell. (2020). https://doi.org/10.1007/s10489-020-01943-6.
Das D, Santosh KC, Pal U. Truncated inception net: COVID-19 outbreak screening using chest X-rays. PhysEngSci Med. (2020);43:915–25. https://doi.org/10.1007/s13246-020-00888-x.
Khan AI, Shah JL, Bhat M. CoroNet: a deep neural network for detection and diagnosis of COVID-19 from chest X-ray images. Comput Methods Progr Biomed. (2020). https://doi.org/10.1016/j.cmpb.2020.105581.
Chollet F. Xception: deep learning with depthwise separable convolutions. In: Proceedings—30th IEEE conference on computer vision and pattern recognition, CVPR 2017. (2017). https://doi.org/10.1109/CVPR.2017.195.
Alom MdZ, Rahman S, Nasrin S, Taha TM,Asari VK. COVID_MTNet: COVID-19 detection with multi-task deep learning approaches. (2020). http://arxiv.org/abs/2004.03747v3 [eess. IV]
Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y. Generative adversarial nets. In: Advances in neural information processing systems. (2014) Cambridge: MIT Press.
Zhu JY, Park T, Isola P, Efros AA. Unpaired image-to-image translation using cycle-consistent adversarial networks. In: Proceedings of the IEEE international conference on computer vision. (2017). https://doi.org/10.1109/ICCV.2017.244
Zebin T, Rezvy S. COVID-19 detection and disease progression visualization: deep learning on chest X-rays for classification and coarse localization. ApplIntell. (2020). https://doi.org/10.1007/s10489-020-01867-1.
Shinde GR, Kalamkar AB, Mahalle PN, et al. Forecasting models for coronavirus disease (COVID-19): a survey of the state-of-the-art. SN ComputSci. (2020);1:197. https://doi.org/10.1007/s42979-020-00209-9.
Joshi A, Dey N, Santosh KC. Intelligent systems and methods to combat COVID-19: Springer briefs in computational intelligence. Cham: Springer; (2020). https://doi.org/10.1007/978-981-15-6572-4 (eBook ISBN: 978-981-15-6572-4).
Santosh KC, Joshi A. COVID-19: prediction, decision-making, and its impacts, book series in lecture notes on data engineering and communications technologies. Berlin: Springer; (2020). https://doi.org/10.1007/978-981-15-9682-7
Sarkar A, Vandenhirtz J, Nagy J, Bacsa D, Riley M. Identification of images of COVID-19 from chest computed tomography (CT) images using deep learning: comparing COGNEX VisionPro Deep Learning 1.0 software with open source convolutional neural networks. (2020). http://arxiv.org/abs/2010.00958v2 [eess.IV]
Acknowledgements
We would like to thank COGNEX for providing their latest Deep Learning software for testing, and the University of Waterloo, along with Darwin AI for collecting and merging the X-ray images from various sources and for providing the python scripts for generating the COVIDx dataset.
Funding
COGNEX funded the project.
Author information
Authors and Affiliations
Contributions
AS wrote the manuscript. AS, JV, JN and DB conducted the experiment. AS, JV, JN, DB, MR analysed the results. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Arjun Sarkar, Joerg Vandenhirtz, Jozsef Nagy, David Bacsa, Mitchell Riley are affiliated with COGNEX.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sarkar, A., Vandenhirtz, J., Nagy, J. et al. Identification of Images of COVID-19 from Chest X-rays Using Deep Learning: Comparing COGNEX VisionPro Deep Learning 1.0™ Software with Open Source Convolutional Neural Networks. SN COMPUT. SCI. 2, 130 (2021). https://doi.org/10.1007/s42979-021-00496-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-021-00496-w