Abstract
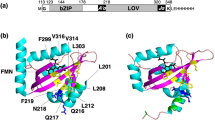
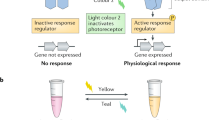
A variety of living organisms including bacteria, fungi, animals, and plants use blue light (BL) to adapt to changing ambient light. Photosynthetic forms (plants and algae) require energy of light for photosynthesis, movements, development, and regulation of activity. Several complex light-sensitive systems evolved in eukaryotic cells to use the information of light efficiently with photoreceptors selectively absorbing various segments of the solar spectrum, being the first components in the light signal transduction chain. They are most diverse in algae. Photosynthetic stramenopiles, which received chloroplasts from red algae during secondary symbiosis, play an important role in ecosystems and aquaculture, being primary producers. These taxa acquired the ability to use BL for regulation of such processes as phototropism, chloroplast photo-relocation movement, and photomorphogenesis. A new type of BL receptor–aureochrome (AUREO)–was identified in Vaucheria frigida in 2007. AUREO consists of two domains: bZIP (basic-region leucine zipper) domain and LOV (light-oxygen-voltage-sensing) domain, and thus this photoreceptor is a BL-sensitive transcription factor. This review presents current data on the structure, mechanisms of action, and biochemical features of aureochromes.
Similar content being viewed by others
Abbreviations
- bHLH:
-
base helix-loop-helix
- BL:
-
blue light
- BLUF:
-
blue light sensing using FAD
- bZIP:
-
basic-region leucine zipper
- LOV:
-
light-oxygen-voltage-sensing
- LRE:
-
light response elements
References
Kianianmomeni, A., and Hallmann, A. (2014) Algal pho-toreceptors: in vivo functions and potential applications, Planta, 239, 1–26.
Ziegler, T., and Moglich, A. (2015) Photoreceptor engineer-ing, Front. Mol. Biosci., 2, doi: 10.3389/fmolb.2015.00030.
Kianianmomeni, A., and Hallmann, A. (2016) Algal pho-tobiology: a rich source of unusual light sensitive proteins for synthetic biology and optogenetics, Methods Mol. Biol., 1408, 37–54.
Fraikin, G. Y., Strakhovskaya, M. G., and Rubin, A. B. (2013) Biological photoreceptors of light dependent regula-tory processes, Biochemistry (Moscow), 78, 1238–1253.
Chattopadhyay, S., Ang, L. H., Puente, P., Deng, X. W., and Wei, N. (1998) Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression, Plant Cell, 10, 673–683.
Luo, X. M., Lin, W. H., Zhu, S., Zhu, J. Y., Sun, Y., Fan, X. Y., Cheng, M., Hao, Y., Oh, E., Tian, M., Liu, L., Zhang, M., Xie, Q., Chong, K., and Wang, Z. Y. (2010) Integration of light-and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis, Dev. Cell, 19, 872–883.
Li, J., Li G., Wang, H., and Deng, X. W. (2011) Phytochrome signaling mechanisms, Arabidopsis Book, 9, e0148; doi: 10.1199/tab.0148.
Ahmad, M., and Cashmore, A. R. (1993) HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor, Nature, 366, 162–166.
Lin, C., and Shalitin, D. (2003) Cryptochrome structure and signal transduction, Annu. Rev. Plant Biol., 54, 469–496.
Mei, Q., and Dvornyk, V. (2015) Evolutionary history of the photolyase/cryptochrome superfamily in eukaryotes, PLoS One, 10, e0135940; doi: 10.1371/journal.pone. 0135940.
Yurina, N. P., Mokerova, D. V., and Odintsova, M. S. (2013) Light-inducible stress plastid proteins of pho-totrophs, Russ. J. Plant Physiol., 60, 577–588.
Kami, C., Lorrain, S., Hornitschek, P., and Fankhauser, C. (2010) Light-regulated plant growth and development, Curr. Top. Dev. Biol., 91, 29–66.
Huala, E., Oeller, P. W., Liscum, E., Han, I. S., Larsen, E., and Briggs, W. R. (1997) Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain, Science, 278, 2120–2123.
Christie, J. M. (2007) Phototropin blue-light receptors, Annu. Rev. Plant Biol., 58, 21–45.
Suetsugu, N., Mittmann, F., Wagner, G., Hughes, J., and Wada, M. (2005) A chimeric photoreceptor gene, NEOCHROME, has arisen twice during plant evolution, Proc. Natl. Acad. Sci. USA, 102, 13705–13709.
Hegemann, P., Fuhrmann, M., and Kateriya, S. (2001) Algal sensory photoreceptors, J. Phycol., 37, 668–676.
Foster, K. W., Saranak, J., Patel, N., Zarilli, G., Okabe, M., Kline, T., and Nakanishi, K. (1984) A rhodopsin is the functional photoreceptor for phototaxis in the unicellular eukaryote Chlamydomonas, Nature, 311, 756–759.
Foster, K. W., and Smyth, R. D. (1980) Light antennas in phototactic algae, Microbiol. Rev., 44, 572–630.
Nagel, G., Ollig, D., Fuhrmann, M., Kateriya, S., Musti, A. M., Bamberg, E., and Hegemann, P. (2002) Channelrhodopsin-1: a light-gated proton channel in green algae, Science, 296, 2395–2398.
Nagel, G., Szellas, T., Huhn, W., Kateriya, S., Adeishvili, N., Berthold, P., Ollig, D., Hegemann, P., and Bamberg, E. (2003) Channelrhodopsin-2, a directly light-gated cation-selective membrane channel, Proc. Natl. Acad. Sci. USA, 100, 13940–13945.
Sineshchekov, O. A., Jung, K. H., and Spudich, J. L. (2002) Two rhodopsins mediate phototaxis to low-and high-inten-sity light in Chlamydomonas reinhardtii, Proc. Natl. Acad. Sci. USA, 99, 8689–8694.
Kato, H. E., Zhang, F., Yizhar, O., Ramakrishnan, C., Nishizawa, T., Hirata, K., Ito, J., Aita, Y., Tsukazaki, T., Hayashi, S., Hegemann, P., Maturana, A. D., Ishitani, R., Deisseroth, K., and Nureki, O. (2012) Crystal structure of the channelrhodopsin light-gated cation channel, Nature, 482, 369–374.
Gomelsky, M., and Klug, G. (2002) BLUF: a novel FAD-binding domain involved in sensory transduction in microorganisms, Trends Biochem. Sci., 27, 497–500.
Gomelsky, M., and Hoff, W. D. (2011) Light helps bacteria make important lifestyle decisions, Trends Microbiol., 19, 441–448.
Fraikin, G. Ya., Strakhovskaya, M. G., Belenikina, N. S., and Rubin, A. B. (2016) LOV and BLUF flavoprotein reg-ulatory photoreceptors of microorganisms and photosenso-ry actuators in optogenetic systems, Moscow Univ. Biol. Sci. Bull., 71, 50–57.
Fiedler, B., Borner, T., and Wilde, A. (2005) Phototaxis in the cyanobacterium Synechocystis sp. PCC 6803: role of different photoreceptors, Photochem. Photobiol., 81, 1481–1488.
Masuda, S., and Bauer, C. E. (2002) AppA is a blue light photoreceptor that anti-represses photosynthesis gene expression in Rhodobacter sphaeroides, Cell, 110, 613–623.
Tschowri, N., Busse, S., and Hengge, R. (2009) The BLUFEAL protein YcgF acts as a direct anti-repressor in a blue-light response of Escherichia coli, Genes Dev., 23, 522–534.
Mussi, M. A., Gaddy, J. A., Cabruja, M., Arivett, B. A., Viale, A. M., Rasia, R., and Actis, L. A. (2010) The oppor-tunistic human pathogen Acinetobacter baumannii senses and responds to light, J. Bacteriol., 192, 6336–6345.
Takahashi, F., Yamagata, D., Ishikawa, M., Fukamatsu, Y., Ogura, Y., Kasahara, M., Kiyosue, T., Kikuyama, M., Wada, M., and Kataoka, H. (2007) AUREOCHROME, a photoreceptor required for photomorphogenesis in stra-menopiles, Proc. Natl. Acad. Sci. USA, 104, 19625–19630.
Ishikawa, M., Takahashi, F., Nozaki, H., Nagasato, C., Motomura, T., and Kataoka, H. (2009) Distribution and phylogeny of the blue light receptors aureochromes in eukaryotes, Planta, 230, 543–552.
Takahashi, F., Hishinuma, T., and Kataoka, H. (2001) Blue light-induced branching in Vaucheria. Requirement of nuclear accumulation in the irradiated region, Plant Cell. Physiol., 42, 274–285.
Banerjee, A., Herman, E., Serif, M., Maestre-Reyna, M., Hepp, S., Pokorny, R., Kroth, P. G., Essen, L. O., and Kottke, T. (2016) Allosteric communication between DNA-binding and light-responsive domains of diatom class I aureochromes, Nucleic Acids Res., 44, 5957–5970.
Serif, M., Lepetit, B., Weißert, K., Kroth, P. G., and Rio Bartulos, C. (2017) A fast and reliable strategy to generate TALEN-mediated gene knockouts in the diatom Phaeodactylum tricornutum, Algal Res., 23, 186–195.
Herman, E., and Kottke, T. (2015) Allosterically regulated unfolding of the A′α helix exposes the dimerization site of the blue-light-sensing aureochrome-LOV domain, Biochemistry, 54, 1484–1492.
Hisatomi, O., Nakatani, Y., Takeuchi, K., Takahashi, F., and Kataoka, H. (2014) Blue light-induced dimerization of monomeric aureochrome-1 enhances its affinity for the tar-get sequence, J. Biol. Chem., 289, 17379–17391.
Conrad, K. S., Manahan, C. C., and Crane, B. R. (2014) Photochemistry of flavoprotein light sensors, Nat. Chem. Biol., 10, 801–809.
Herrou, J., and Crosson, S. (2011) Function, structure and mechanism of bacterial photosensory LOV proteins, Nat. Rev. Microbiol., 9, 713–723.
Harper, S. M., Neil, L. C., and Gardner, K. H. (2003) Structural basis of a phototropin light switch, Science, 301, 1541–1544.
Moglich, A., and Moffat, K. (2007) Structural basis for light-dependent signaling in the dimeric LOV domain of the photosensor YtvA, J. Mol. Biol., 373, 112–126.
Nash, A. I., McNulty, R., Shillito, M. E., Swartz, T. E., Bogomolni, R. A., Luecke, H., and Gardner, K. H. (2011) Structural basis of photosensitivity in a bacterial light-oxy-gen-voltage/helix-turn-helix (LOV-HTH) DNA-binding protein, Proc. Natl. Acad. Sci. USA, 108, 9449–9454.
Vaidya, A. T., Chen, C. H., Dunlap, J. C., Loros, J. J., and Crane, B. R. (2011) Structure of a light-activated LOV pro-tein dimer that regulates transcription, Sci. Signal., 4, ra50; doi: 10.1126/scisignal.2001945.
Rodriguez-Romero, J., Hedtke, M., Kastner, C., Muller, S., and Fischer, R. (2010) Fungi, hidden in soil or up in the air: light makes a difference, Annu. Rev. Microbiol., 64, 585–610.
Schwerdtfeger, C., and Linden, H. (2003) VIVID is a flavo-protein and serves as a fungal blue light photoreceptor for photoadaptation, EMBO J., 22, 4846–4855.
Green, J., Crack, J. C., Thomson, A. J., and LeBrun, N. E. (2009) Bacterial sensors of oxygen, Curr. Opin. Microbiol., 12, 145–151.
Banerjee, A., Herman, E., Kottke, T., and Essen, L. O. (2016) Structure of a native-like aureochrome 1a LOV domain dimer from Phaeodactylum tricornutum, Structure, 24, 171–178.
Salomon, M., Christie, J. M., Knieb, E., Lempert, U., and Briggs, W. R. (2000) Photochemical and mutational analy-sis of the FMN-binding domain of the plant blue light receptor, phototropin, Biochemistry, 39, 9401–9410.
Heintz, U., and Schlichting, I. (2016) Blue light-induced LOV domain dimerization enhances the affinity of aure-ochrome 1a for its target DNA sequence, eLife, 5, e11860; doi: 10.7554/eLife.11860.
Mitra, D., Yang, X., and Moffat, K. (2012) Crystal struc-tures of aureochrome1 LOV suggest new design strategies for optogenetics, Structure, 20, 698–706.
Essen, L. O., Franz, S., and Banerjee, A. (2017) Structural and evolutionary aspects of algal blue light receptors of the cryptochrome and aureochrome type, J. Plant Physiol., 217, 27–37.
Chen, Z., Yang, M. K., Li, C. Y., Wang, Y., Zhang, J., Wang, D. B., Zhang, X. E., and Ge, F. (2014) Phosphoproteomic analysis provides novel insights into stress responses in Phaeodactylum tricornutum, a model diatom, J. Proteome Res., 13, 2511–2523.
Herman, E., Sachse, M., Kroth, P. G., and Kottke, T. (2013) Blue-light-induced unfolding of the Jα helix allows for the dimerization of aureochrome-LOV from the diatom Phaeodactylum tricornutum, Biochemistry, 52, 3094–3101.
Harper, S. M., Christie, J. M., and Gardner, K. H. (2004) Disruption of the LOV–Jα helix interaction activates pho-totropin kinase activity, Biochemistry, 43, 16184–16192.
Kroth, P. G., Wilhelm, C., and Kottke, T. (2017) An update on aureochromes: phylogeny–mechanism–function, J. Plant Physiol., 217, 20–26.
Akiyama, Y., Nakasone, Y., Nakatani, Y., Hisatomi, O., and Terazima, M. (2016) Time-resolved detection of light-induced dimerization of monomeric aureochrome-1 and change in affinity for DNA, J. Phys. Chem., 120, 7360–7370.
Toyooka, T., Hisatomi, O., Takahashi, F., Kataoka, H., and Terazima, M. (2011) Photoreactions of aureochrome-1, Biophys. J., 100, 2801–2809.
Takahashi, F. (2016) Blue-light-regulated transcription fac-tor, Aureochrome, in photosynthetic stramenopiles, J. Plant Res., 129, 189–197.
Hisatomi, O., Terauchi, K., Zikihara, K., Ookubo, Y., Nakatani, Y., Takahashi, F., Tokutomi, S., and Kataoka, H. (2013) Blue light-induced conformational changes in a light-regulated transcription factor, Aureochrome-1, Plant Cell. Physiol., 54, 93–106.
Deng, Y., Yao, J., Fu, G., Guo, H., and Duan, D. (2014) Isolation, expression, and characterization of blue light recep-tor AUREOCHROME gene from Saccharina japonica (Laminariales, Phaeophyceae), Mar. Biotechnol., 16, 135–143.
Ishikawa, M., Kataoka, H., and Takahashi, F. (2012) Analysis of light-dependent cell morphology and an accumulation response in Ochromonas danica, Cytologia, 77, 465–473.
Huysman, M. J., Fortunato, A. E., Matthijs, M., Costa, B. S., Vanderhaeghen, R., Van den Daele, H., Sachse, M., Inze, D., Bowler, C., Kroth, P. G., Wilhelm, C., Falciatore, A., Vyverman, W., and De Veylder, L. (2013) Aureochome1a-mediated induction of the diatom-specific cyclin dsCYC2 controls the onset of cell division in Diatoms (Phaeodactylum tricornutum), Plant Cell., 25, 215–228.
Schellenberger Costa, B., Sachse, M., Jungandreas, A., Bartulos, C. R., Gruber, A., Jakob, T., Kroth, P. G., and Wilhelm, C. (2013) Aureochrome 1a is involved in the pho-toacclimation of the Diatom Phaeodactylum tricornutum, PLoS One, 8, e74451.
Glantz, S. T., Carpenter, E. J., Melkonian, M., Gardner, K. H., Boyden, E. S., Wong, G. K.-S., and Chow, B. Y. (2016) Functional and topological diversity of LOV domain pho-toreceptors, Proc. Natl. Acad. Sci. USA, 113, 1442–1451.
Archibald, J. M. (2015) Endosymbiosis and eukaryotic cell evolution, Curr. Biol., 25, 911–921.
Di Roberto, R. B., and Peisajovich, S. G. (2014) The role of domain shuffling in the evolution of signaling networks, J. Exp. Zool. B Mol. Dev. Evol., 322, 65–72.
Li, F.-W., Rothfels, C. J., Melkonian, M., Villarreal, J. C., Stevenson, D. W., Graham, S. W., Wong, G. K. S., Mathews, S., and Pryer, K. M. (2015) The origin and evo-lution of phototropins, Front. Plant Sci., 6, 637.
Janouskovec, J., Horak, A., Obornik, M., Lukes, J., and Keeling, P. J. (2010) A common red algal origin of the api-complexan, dinoflagellate, and heterokont plastids, Proc. Natl. Acad. Sci. USA, 107, 10949–10954.
Suetsugu, N., and Wada, M. (2013) Evolution of three LOV blue light receptor families in green plants and photosyn-thetic stramenopiles: phototropin, ZTL/FKF1/LKP2 and aureochrome, Plant Cell Physiol., 54, 8–23.
Kasahara, M., Torii, M., Fujita, A., and Tainaka, K. (2010) FMN binding and photochemical properties of plant puta-tive photoreceptors containing two LOV domains, LOV/LOV proteins, J. Biol. Chem., 285, 34765–34772.
Kagawa, T., Kasahara, M., Abe, T., Yoshida, S., and Wada, M. (2004) Function analysis of phototropin2 using fern mutants deficient in blue light-induced chloroplast avoid-ance movement, Plant Cell. Physiol., 45, 416–426.
Grusch, M., Schelch, K., Riedler, R., Reichhart, E., Differ, C., Berger, W., Ingles-Prieto, A., and Janovjak, H. (2014) Spatiotemporally precise activation of engineered receptor tyrosine kinases by light, EMBO J., 33, 1713–1726.
Muller, K., and Weber, W. (2013) Optogenetic tools for mammalian systems, Mol. BioSyst., 9, 596–608.
Pathak, G. P., Vrana, J. D., and Tucker, C. L. (2013) Optogenetic control of cell function using engineered pho-toreceptors, Biol. Cell, 105, 59–72.
Ye, H., Daoud-El Baba, M., Peng, R. W., and Fussenegger, M. (2011) A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice, Science, 332, 1565–1568.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © A. B. Matiiv, E. M. Chekunova, 2018, published in Biokhimiya, 2018, Vol. 83, No. 6, pp. 839-851.
Rights and permissions
About this article
Cite this article
Matiiv, A.B., Chekunova, E.M. Aureochromes – Blue Light Receptors. Biochemistry Moscow 83, 662–673 (2018). https://doi.org/10.1134/S0006297918060044
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297918060044