Abstract
Deep neural networks have been used in survival prediction by providing high-quality features. However, few works have noticed the significant role of topological features of whole slide pathological images (WSI). Learning topological features on WSIs requires dense computations. Besides, the optimal topological representation of WSIs is still ambiguous. Moreover, how to fully utilize the topological features of WSI in survival prediction is an open question. Therefore, we propose to model WSI as graph and then develop a graph convolutional neural network (graph CNN) with attention learning that better serves the survival prediction by rendering the optimal graph representations of WSIs. Extensive experiments on real lung and brain carcinoma WSIs have demonstrated its effectiveness.
This work was partially supported by NSF IIS-1423056, CMMI-1434401, CNS-1405985, IIS-1718853 and the NSF CAREER grant IIS-1553687.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
- Graph Convolutional Neural Networks (GCN)
- Pathological Images
- Survival Prediction
- Optimal Topological Representation
- National Lung Screening Trial (NLST)
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Survival analysis is generally a set of statistical models where the output is the elapsed time until a certain event occurs. The event can range from vehicle part failure to adverse drug reaction. Clinical trials are aimed to assess different treatment regimes with the biological death as primary event of interest to observe. An accurate estimate of survival probability provides invaluable information for clinical interventions.
The Cox proportional hazards model [3] is most popular in survival analysis. However, the classical Cox model and its early followers overly simplified the patient’s survival probability as linear mapping from covariates. Recently, Katzman et al. designed a fully connected network (DeepSurv [9]) to learn the nonlinear survival functions. Although it was showed that neural networks outperformed the linear Cox model [4], it cannot directly learn from pathological images. Along with the success of convolutional neural networks (CNNs) on generic images, pathological image, as well as CT and MRI [14], have become ideal data sources for training DL-based survival models. Among them, whole slide image (WSI) [12] is one of the most valuable data formats due to the massive multi-level pathological information on nidus and its surrounding tissues.
WSISA [21] was the first trial of moving survival prediction onto whole slide pathological images. To have a efficient approach on WSIs, a patch sampling on WSIs is inevitable. However, their DeepConvSurv model was trained on clustered patch samples separately. Consequently, the features extracted were over-localized for WSIs because the receptive field is constrained within physical area corresponding to a single patch (0.063 mm\(^2\)). The pathological sections of nidus from patients contain more than the regions of interest (e.g tumor cells), therefore, the representations from random patch may not strongly correspond to the disease. Furthermore, it has been widely recognized that the topological properties of instances on pathological images are crucial in medical tasks, e.g. cell subtype classification and cancer classification. While, WSISA is neither able to learn global topological representations of WSIs nor to construct feature maps upon given topological structures.
Graph is widely employed to represent topological structures. However, modeling a WSI as graph is not straightforward. Cell-graph [6] is infeasible for WSIs due to its huge number of cells and the many possible noisy nodes (isolated cells). The intermediate patch-wise features are a good option to construct graph with, balancing efficiency and granularity. However, applying CNNs on graph-structured data is still difficult.
In the paper, we propose a graph convolutional neural network (GCN) based survival analysis model (DeepGraphSurv) where global topological features of WSI and local patch features are naturally integrated via spectral graph convolution operators. The contributions are summarized as: (1) learn both local and global representations of WSIs simultaneously: local patch features are integrated with global topological structures through convolution; (2) task-driven adaptive graphs induce better representations of WSI; (3) introducing graph attention mechanism reduces randomness of patch sampling and therefore increases model robustness. As far as we know, DeepGraphSurv is the first GCN based survival prediction model with WSIs as input. Extensive experiments on cancer patient WSI datasets demonstrate that our model outperforms the state-of-the-art models by providing more accurate survival risk predictions.
2 Methodology
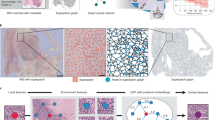
Graph Construction on WSI: Given a set of sampled patch images \(\mathbf {P}=\{P_i\}\) from WSI, we have to dump those patches from the margin areas which contains few cells, therefore, the cardinality \(\Vert \mathbf {P}\Vert \) differs by WSI. Consequently, the graphs we construct for WSIs are of different sizes. Given patches as vertices, vertex features are generated by the VGG-16 network pre-trained on ImageNet. Due to the lack of patch labels, we cannot fine-tune the network on WSI patches. We will introduce how graph CNN model mitigates this deficiency in next section. Graph edges were constructed by thresholding the Euclidean distances between patch pairs, which were calculated using the 128 features compressed from the VGG-16 outputs with patches as input. Compressions are committed separately on train and test sets by principal component analysis (PCA).
Spectral Graph Convolution. Given a graph \(\mathcal {G}=(V, E)\), its normalized graph Laplacian \(L = I - D^{-1/2} A D^{-1/2}\), where A is the adjacency matrix and D is the degree matrix of \(\mathcal {G}\). The graph on WSI is irregular with \(\varDelta (\mathcal {G})\gg \delta (\mathcal {G})\). A spectral convolutional filter built based on spectral graph theory [1, 2, 20] is more applicable to irregular WSI graph. It was proved that a spectrum formed by smooth frequency components leads to a localized spatial kernel. Furthermore, [5] formulated kernel as a \(K^{th}\) order polynomial of diagonal \(\varLambda \), and \(diag(\varLambda )\) is the spectrum of graph Laplacian L:
Based on theorem from [2], spectral convolution on graph \(\mathcal {G}\) with vertex features \(X\in \mathrm {R}^{N\times F}\) as layer input is formulated as:
ReLU is activation function. Output \(Y\in \mathrm {R}^{N\times F}\) is a graph of identical number of vertices with convolved features. The learning complexity of K-localized kernel is \(\mathcal {O}(K)\). To have a fast filtering, [5] used Chebyshev expansion as approximation of \(g_{\theta }(L)\), Recursive calculation of \(g_{\theta }(L^K)\) reduces the time cost from \(\mathcal {O}(K N^2)\) to \(\mathcal {O}(KS)\), S (\(\ll N^2\)) is the count of nonzeros in L. Sparseness of L was enforced by edge thresholding when graph construction. Initial WSI graph \(\mathcal {G}\) was built upon compressed patch features. The VGG-16 feature network was not fine-tuned on WSI patches due to the lack of patch labels. Patient-wise censored survival label is absolutely infeasible for a patch-wise training. Therefore, the initial graph may not correctly represent the topological structures between patches on WSI.
Survival-Specific Graph. The deficiency of initial graph results from the insufficiently trained feature network. It has two problems: (1) network used irrelevant supervision (i.e ImageNet label); (2) network was not fine-tuned on pathological images. It would be better, if the patch features could be fine-tuned with survival censor labels. To achieve it, we design a separate graph \(\tilde{\mathcal {G}}\) and \(\tilde{L}\) to describe the specific survival-related topological relationship between WSI patches [13, 15]. \(\tilde{L}\) is learned individually on each WSI. Direct learning of \(\tilde{L}\) is impractical because of the graph size and the uniqueness of topology on WSIs. Instead of learning and storing graph edges, we learn the Mahalanobis distance metrics M for evaluating edge connectivity. If d is the dimensionality of feature, the learning complexity is reduced from \(\mathcal {O}(N^2)\) to \(\mathcal {O}(d^2)\). Because there is no priors on metrics, M has to be randomly initialized. To accelerate the convergence, we keep initial graph as regularization term for survival-specific graph. The final graph Laplacian in convolution will be \(\mathcal {L}(M, X) = \tilde{L}(M,X)+\beta L \). \(\beta \) is trade-off coefficient. With survival-specific graph, the proposed graph convolution is formulated as:
Afterwards, there is a feature transform operator parameterized as \(W\in \mathrm {R}^{F_{in}\times F_{out}}\) and bias \(b\in \mathrm {R}^{F_{out}}\) applied to output Y: \(Y'=Y W + b\). This re-parameterization on activations will lead to a better imitation of CNNs, whose output features are mappings of all input feature dimensions. Model parameters \(\{M, \theta \}\) get updated by back-propagation w.r.t. survival loss, which promises fine-tuned features and graphs optimized for survival analysis purpose.
Graph Attention Mechanism. Generally, there are merely a few local regions of interest (RoIs) on WSIs matter in survival analysis. Random sampling cannot guarantee patches are all from RoIs. Attention mechanism provides an adaptive patch selection by learning “importance” on them. In DeepGraphSurv, there is a parallel network to learn attention on nodes conditioned on node features. The network consists of two proposed GCN layers (Eq. 3). The outputs of attention network are node attention values: \(\alpha =f_{attn}'(X)\). Given learned attentions, the output risk R for X on graph \(\mathcal {G}(V,E)\) is the weighted sum of \(Y_n\) of each node n:
As shown above, in graph gather layer (Fig. 1), the learned attentions are multiplied onto the node-wise predictions when aggregating attentive graph outputs. The attention network will be trained jointly with the prediction network. Different from previous DL-based survival models that basically act as feature extractor [21], DeepGraphSurv directly generates predicted risks. We integrated regression of survival risk with graph feature learning on WSIs. The loss function is negative Cox log partial likelihood for censored survival data:
\(S_i\), \(T_i\) are respectively the censor status and the survival time of i-th patient. The fine-tuned patch features and the survival-specific graphs of WSIs are accessible at each proposed GCN layer, while the later layers offer more high-level topology-aware features of WSI.
3 Experiment
3.1 Dataset
As to the raw data source, we utilized the whole slide pathological images from a generic cancer patient dataset TCGA, publicly released by The Cancer Genome Atlas project [8]. The research studied what and how errors in DNA trigger the occurrence of 33 cancer subtypes. We tested our model on two cancer subtypes from TCGA data: glioblastoma multiforme (GBM) and lung squamous cell carcinoma (LUSC). Besides, NLST (National Lung Screening Trials [10]) employed 53,454 heavy smokers of age 55 to 74 with at least 30-year smoking history as high risk group for lung cancer survival analysis. We also committed an experiment on a subset of NLST database that consists of both squamous-cell carcinoma (SCC) and adenocarcinoma (ADC) patients’ WSIs to evaluate the performance of our model on mixed cancer subtype dataset. Some quantitative facts of WSI used in the experiments are listed in Table 1.
3.2 State-of-the-Art Methods
The baseline survival methods include: LASSO-Cox model [18], BoostCI [17] and Multi-Task Learning model for Survival Analysis (MTLSA) [16]. However, their effectiveness largely depends on the quality of hand-crafted features. Moreover, they were entirely not designed for WSI based survival analysis. For a fair comparison, we first feed those models with the features extracted by CellProfiler [11], e.g cell shape and textures, sampled and averaged over patch images. Then, we feed them with the WSI features generated by DeepGraphSurv from the same group of patient in order to demonstrate the gain of performance brought by the fine-tuned topology-aware WSI features only.
Besides classical models, we compared DeepGraphSurv with the state-of-the-art deep learning based survival models on WSI. WSISA [21] worked on clustered patches from WSIs, however, they simply neglected the topological relationship of the instances on WSI, which is also of great importance on survival analysis. Graph CNNs have recognized power of mining structured features on graph data. We concatenate the latest spectral GCN model [5], working on pre-trained fixed graphs, with a Cox regression as one of comparison methods in order to confirm the advantages brought by adding proposed survival-specific graphs onto GCN.
3.3 Result and Discussion
As far as we know, DeepGraphSurv is the first survival model that used attention scheme. Figure 2 shows that, after 40 epoch, the regions of high attention on a WSI have correctly highlighted the most of RoIs annotated by medical experts. This interpreted part of global structural knowledge we have discovered on WSI.
The concordance probability (C-index) is the fraction of all pairs of patients whose predicted survival times are correctly ordered as all censored patients that can be reasonably ordered. Forming survival order as graph \(\mathcal {G}_t(\mathcal {D}, \mathcal {E})\) whose edge \(\mathcal {E}_{i,j}\) implies \(T_i < T_{j}\), the C-index is: \( C(\mathcal {D},\mathcal {G}_t, f(x)) = \frac{1}{\Vert \mathcal {E} \Vert } \sum _{\mathcal {E}_{i,j}} \mathbf {1}_{f(x_i) < f(x_j)}\), where \(\mathbf {1}_{f(x_i) < f(x_j)}\) is the indicator function: \(\mathbf {1}_{a<b}=1\) if \(a<b\), otherwise 0. \(f(x_i)\) is the predicted risk of \(x_i\). When a patient has multiple WSIs, the predicted risks were first averaged for the patient before calculating C-index.
The C-index results are reported in Table 2. Training and testing sets were randomly splitted and separately prepared. The classical survival models, e.g LASSO-Cox, cannot perform well was because they only utilize hand-crafted features. Possible issues include: (1) patches are partial representations of WSI; (2) the data quality of patch may vary. Consequently, the features collected from random patches brought noisy and biased representations of WSI. Moreover, the features from CellProfiler are general descriptors of pathological images. After feeding them with the WSI features generated by DeepGraphSurv, the C-index were largely lifted by 0.04 on average on NLST and LUSC. This outcome showed that the features fine-tuned with survival labels are indeed better representations of WSI for survival analysis purpose.
However, we also observe that, due to the lower image quality, only using fine-tuned patch features cannot improve prediction on GBM data. DeepGraphSurv generates predictions by encoding patch features with their topological structure via convolution. When patch features are unreliable, topological structure of WSI instances makes more sense in recognition of survival patterns. This may explain the lift by DeepGraphSurv compared to [5, 21] who learn little from topology.
The previous GCN [5] outperformed WSISA [21] on most of datasets because it can aggregate node features as graph representation of WSI according to graph structure, while [21] cannot. However, [5] still worked on unsupervised graphs obtained with noisy VGG-16 features. DeepGraphSurv conducted convolution on the fine-tuned survival-specific graphs that were trained to represent the survival-related topological structures on each individual WSI. This improved C-index by another 0.03 on average, which again verified that the topological features trained in supervised way work better than that learned from unsupervised approaches.
4 Conclusion
Survival prediction is a useful clinical intervention tool, although it cannot act as expected in many scenarios. Efficient mining of survival-related structured features on whole slide images is a promising solution of boosting survival analysis. In this paper, we suggested to model WSI as graph and proposed DeepGraphSurv to learn global topological representations of WSI. Instead of unsupervised graph, DeepGraphSurv creatively utilized a survival-specific graph trained under supervision of survival labels. The effectiveness of our model has been confirmed by improved accuracy of risk ranking on multiple cancer patient datasets across carcinoma subtypes.
References
Chen, P.Y., Zhang, B., Al Hasan, M.: Incremental eigenpair computation for graph laplacian matrices: theory and applications. Soc. Netw. Anal. Min. 8(1), 4 (2018)
Chung, F.R.: Spectral Graph Theory, no. 92. American Mathematical Society, Providence (1997)
Cox, D.R.: Regression models and life-tables. J. R. Stat. Society. Ser. B (Methodological) 34, 187–220 (1972)
Dave, V.S., Al Hasan, M., Zhang, B., Reddy, C.K.: Predicting interval time for reciprocal link creation using survival analysis. Soc. Netw. Anal. Min. 8(1), 16 (2018)
Defferrard, M., Bresson, X., Vandergheynst, P.: Convolutional neural networks on graphs with fast localized spectral filtering. In: Advances in Neural Information Processing Systems, pp. 3837–3845 (2016)
Gunduz, C., Yener, B., Gultekin, S.H.: The cell graphs of cancer. Bioinformatics 20(suppl\(\_\)1), i145–i151 (2004)
Kalbfleisch, J.D., Prentice, R.L.: The Statistical Analysis of Failure Time Data, vol. 360. Wiley, Hoboken (2011)
Kandoth, C., et al.: Mutational landscape and significance across 12 major cancer types. Nature 502(7471), 333–339 (2013)
Katzman, J., Shaham, U., Cloninger, A., Bates, J., Jiang, T., Kluger, Y.: Deep survival: a deep cox proportional hazards network. arXiv preprint arXiv:1606.00931 (2016)
Kramer, B.S., Berg, C.D., Aberle, D.R., Prorok, P.C.: Lung cancer screening with low-dose helical CT: results from the national lung screening trial (NLST). J. Med. Screen 18(3), 109–111 (2011)
Lamprecht, M.R., Sabatini, D.M., Carpenter, A.E., et al.: Cellprofiler™: free, versatile software for automated biological image analysis. Biotechniques 42(1), 71 (2007)
Li, R., Huang, J.: Fast regions-of-interest detection in whole slide histopathology images. In: Wu, G., Coupé, P., Zhan, Y., Munsell, B., Rueckert, D. (eds.) Patch-MI 2015. LNCS, vol. 9467, pp. 120–127. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-28194-0_15
Li, R., Huang, J.: Learning graph while training: an evolving graph convolutional neural network. arXiv preprint arXiv:1708.04675 (2017)
Li, R., Li, Y., Fang, R., Zhang, S., Pan, H., Huang, J.: Fast preconditioning for accelerated multi-contrast MRI reconstruction. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9350, pp. 700–707. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24571-3_84
Li, R., Wang, S., Zhu, F., Huang, J.: Adaptive graph convolutional neural networks. arXiv preprint arXiv:1801.03226 (2018)
Li, Y., Wang, J., Ye, J., Reddy, C.K.: A multi-task learning formulation for survival analysis. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 1715–1724 (2016)
Mayr, A., Schmid, M.: Boosting the concordance index for survival data-a unified framework to derive and evaluate biomarker combinations. PloS one 9(1), e84483 (2014)
Tibshirani, R., et al.: The lasso method for variable selection in the cox model. Stat. Med. 16(4), 385–395 (1997)
Yang, Y., Zou, H.: A cocktail algorithm for solving the elastic net penalized cox’s regression in high dimensions. Stat. Interface 6(2), 167–173 (2013)
Zhang, B., Hasan, M.A.: Name disambiguation in anonymized graphs using network embedding. In: Proceedings of the 26th ACM International on Conference on Information and Knowledge Management (2017)
Zhu, X., Yao, J., Zhu, F., Huang, J.: Wsisa: making survival prediction from whole slide histopathological images. In: IEEE Conference on Computer Vision and Pattern Recognition, pp. 7234–7242 (2017)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Li, R., Yao, J., Zhu, X., Li, Y., Huang, J. (2018). Graph CNN for Survival Analysis on Whole Slide Pathological Images. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11071. Springer, Cham. https://doi.org/10.1007/978-3-030-00934-2_20
Download citation
DOI: https://doi.org/10.1007/978-3-030-00934-2_20
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00933-5
Online ISBN: 978-3-030-00934-2
eBook Packages: Computer ScienceComputer Science (R0)