Abstract
Automatic diagnosis of diabetic retinopathy (DR) using retinal fundus images is a challenging problem because images of low grade DR may contain only a few tiny lesions which are difficult to perceive even to human experts. Using annotations in the form of lesion bounding boxes may help solve the problem by deep learning models, but fully annotated samples of this type are usually expensive to obtain. Missing annotated samples (i.e., true lesions but not included in annotations) are noise and can affect learning models negatively. Besides, how to utilize lesion information for identifying DR should be considered carefully because different types of lesions may be used to distinguish different DR grades. In this paper, we propose a new framework for unifying lesion detection and DR identification. Our lesion detection model first determines the missing annotated samples to reduce their impact on the model, and extracts lesion information. Our attention-based network then fuses original images and lesion information to identify DR. Experimental results show that our detection model can considerably reduce the impact of missing annotation and our attention-based network can learn weights between the original images and lesion information for distinguishing different DR grades. Our approach outperforms state-of-the-art methods on two grand challenge retina datasets, EyePACS and Messidor.
Z. Lin, R. Guo, Y. Wang—These authors contributed equally to this work.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
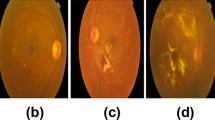
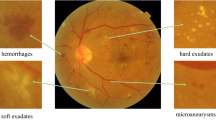
Diabetic retinopathy (DR) is one of the most severe complications of diabetes, which can cause vision loss or even blindness. DR can be identified by ophthalmologists based on the type and count of lesions. Usually, the severity of DR is rated on a scale of 0 to 4: normal, mild, moderate, severe, and proliferative. As shown in Fig. 1(b), grades 1 to 3 are classified as non-proliferative DR (NPDR), which can be identified by the amount of lesions including microaneurysm (MA), hemorrhages (HE), and Exudate (EXU). Grade 4 is proliferative DR (PDR) whose lesions (such as retinal neovascularization (RNV)) are different from those of other grades. Ophthalmologists can identify the presence of DR by examining digital retinal fundus images, but this is a time-consuming and manual-intensive process. Thus, it is important to develop an automatic method to assist DR diagnosis for better efficiency and reducing expert labor.
(a) Missing annotated lesions in images. Yellow dotted boxes are ophthalmologists’ notes and blue arrows indicate missing annotation. (b) DR grades can be identified by the types and count of lesions (yellow: MA, blue: HE, green: EXU, and red: RNV). The lesions for Grade 4 are different from those of other grades.
There are mainly two kinds of machine learning methods for identifying DR. The first kind uses image-level labels to train a classification model that distinguishes DR grades directly. Kumar et al. [7] tackled this task as abnormality detection using a mixture model. Recently, deep learning techniques, such as convolution neural networks (CNN), have been employed to identify DR [5][3]. Wang et al. [11] used CNN feature maps to find the more important locations, thus improving the performance. But, tiny lesions (e.g., MA and HE) may be neglected by these methods with only image-level labels, affecting prediction accuracy, especially for DR grades 1 and 2. The second kind of methods first detects lesions for further processing. Dai et al. [2] tried to detect lesions using clinical reports. van Grinsven et al. [4] sped up model training by selective data sampling for HE detection. Seoud et al. [9] used hand-crafted features to detect retinal lesions and identify DR grade. Yang et al. [13] gave a two-stage framework for both lesion detection and DR grading using annotation of locations including MA, HE, and EXU.
Fusing lesion information to identify DR can effectively help the models perform better. However, there are still other difficulties to handle: (i) A common problem is that usually not all lesions are annotated. In retinal fundus images, the amount of MA and HE is often relatively large, and experts may miss quite some lesions (e.g., see Fig. 1(a)). Note that the missing annotated lesions are treated as negative samples (i.e., background) and thus are “noise” to the model. (ii) Not all kinds of lesions are beneficial to distinguishing all DR grades. For example, DR grade 4 (PDR) can be identified using RNV lesions, but has no direct relationship with MA and HE lesions (see Fig. 1(b)). If we fuse the information of these two types of lesions directly, it may be noisy information to detecting PDR and affect the model’s performance.
To handle these difficulties, we develop a new framework for identifying DR using retinal fundus images based on annotation that includes DR grades and bounding boxes of MA and HE lesions (possibly with a few missing annotated lesions). We first extract lesion information into a lesion map by a detection model, and then fuse it with the original image for DR identification. To deal with noisy negative samples induced by missing annotated lesions, our detection model uses center loss [12], which can cluster the features of similar samples around a feature center called Lesion Center. We also propose a sampling method, called Center-Sample, to find noisy negative samples by measuring their features’ similarity to the Lesion Center and reduce their sampling probabilities. Besides, we adapt center loss from classification tasks to detection tasks efficiently, which makes the model more discriminative and robust. In the classification stage, we integrate feature maps of the original images and lesion maps using an Attention Fusion Network (AFN). AFN can learn the weights between the original images and lesion maps when identifying different DR grades to reduce the interference of unnecessary lesion information on classification. We evaluate our framework using datasets collected from a local hospital and two public datasets, EyePACS and Messidor. Experimental results show that our Center-Sample mechanism can effectively determine noisy samples and achieve promising performance. Our AFN can utilize lesion information well and outperform the state-of-the-art methods on the two public datasets.
The Center-Sample Detector (left) predicts the probabilities of the lesions using the anti-noise Center-Sample Module. Then AFN (right) uses the original image and detection model output as input to identify DR (\(f_{les}\) and \(f_{ori}\) are feature maps, \(W_{les}\) and \(W_{ori}\) are attention weights).
2 Method
This section presents the key components of our approach, the Center-Sample detector and Attention Fusion Network. As shown in Fig. 2, the detection model predicts the probabilities of the lesions in the entire image. Then AFN uses both the original image and detection model output as input to identify DR grades.
2.1 Center-Sample Detector
The Center-Sample detector aims to detect n types of lesions (here, \(n=2\), for MA and HE) in a fundus image. Figure 2 gives an overview of the Center-Sample detector, which composes of three main parts: shared feature extractor, classification/bounding box detecting header, and Noisy Sample Mining module.
The first two parts form the main network for lesion detection to predict the lesion probability map. Their main structures are adapted from SSD [8]. The backbone until conv4_3 is used as feature extractor, and the detect headers are the same as SSD. The third part includes two components: Sample Clustering for clustering similar samples and Noisy Sample Mining for determining the noisy samples and reducing their sampling weight.
Sample Clustering. Here we show how to adapt center loss in classification tasks to detection tasks and how to cluster similar samples using center loss. This component begins by taking the feature map from the shared feature extractor, which is a tensor of size \(h \times w \times c\). We transform it to a feature map u of size \(h \times w \times d\) (\(d \ll c\)) by adding 1\(\times \)1 convolution layers after the shared feature extractor. Each position \(u_{ij}\) in u is a d-D vector, called deep feature, as shown in Fig. 2. That is, \(u_{ij}\) is a feature vector mapped from a corresponding position patch \(f_{ij}\) in the original image to a high-dimensional feature space S, where \(f_{ij}\) denotes the receptive field of \(u_{ij}\). We assign each \(u_{ij}\) with a label indicating whether a lesion is in the corresponding position and (if yes) which type of lesion it is (in Fig. 2, different colors are for different labels). Thus, there are totally \(n+1\) label classes including background (no lesion in corresponding location) and n classes of lesions. We treat background as negative samples and the n classes of lesions as positive samples. Then, we average the deep features \(u_{ij}\) of each class to obtain \(n+1\) feature centers (the centers of positive labels are called lesion centers), and make the \(u_{ij}\) cluster around their corresponding center in the space S using center loss [12] (in Fig. 2, the triangles denote the centers): \(\mathcal {L}_C = \frac{1}{2} \sum _{i=1}^{w} \sum _{j=1}^{h} ||u_{ij} - c_{y_{ij}}||_2^2\), where \({y_{ij}} \in [0, n]\) is the corresponding label of \(u_{ij}\) in location (i, j), and \(c_{y_{ij}} \in \mathbb {R}^d \) is the center of the \(y_{ij}\)-th class. During the detection training phase, we minimize \(\mathcal {L}_C\) and simultaneously update the feature centers using the SGD algorithm in each iteration, to make the \(u_{ij}\) cluster to the center \(c_{y_{ij}}\). Note that the deep features \(u_{ij}\) of noisy negative samples become closer to the corresponding lesion center \(c_{y_{ij}}\) than true negative samples after several iterations.
Noisy Sample Mining. In the Noisy Sample Mining module, we reduce the impact of noisy negative samples by down-weighting them. First, for each \(u_{ij}\) labeled as a negative sample, we select the minimum \(\mathcal {L}2\) distance between \(u_{ij}\) and all lesion centers, denoted by min-\(dist_{ij}\) and sort all elements in min-dist in increasing order. Then, the sampling probability \(P(u_{ij})\) is assigned as:
where \(r_{ij}\) is the rank of \(u_{ij}\) in min-dist. Note that \(u_{ij}\) is close to lesion centers if \(r_{ij}\) is small. The lower bound \(t_l\) and upper bound \(t_u\) of sampling ranking and \(\gamma \) are three hyper-parameters. If \(r_{ij} < t_u\), then \(u_{ij}\) shall be a noisy sample with high probability and we ignore it by setting the sampling probability to 0. \(P(u_{ij})\) is set to 1.0 when \(r_{ij} > t_u\) for treating \(u_{ij}\) as a true negative sample. \(\gamma \) smoothly adjusts the sampling probability between ranks \(t_l\) and \(t_u\). We treat the summation of \(\mathcal {L}_C\) and detection loss in [8] as multi-task loss for robustness. In [12], center loss is required for a comparable large batch size for stable center gradient computing, but in our method, a large number of deep features ensures the stability in small batch size.
During the training phase, we train the model with cropped patches of the original images that include lesions. During the inference phase, a whole image is fed to the trained model, and the output is a tensor M of size \(h \times w \times n\), where every n-D vector \(M_{ij}\) in M denotes the maximum probability among all Anchor Boxes in this position for each lesion. We take this tensor, called Lesion Map, as the input of the Attention Fusion Network.
2.2 Attention Fusion Network
As stated in Sect. 1, some lesion information can be noise to identifying certain DR grades. To resolve this issue, we propose an information fusion method based on attention mechanism [1], called Attention Fusion Network (AFN). AFN can produce the weights based on the original images and lesion maps to reduce the impact of unneeded lesion information for identifying different DR grades. AFN contains two feature extractors and an attention network (see Fig. 2). The scaled original images and lesion maps are the inputs of two separate feature extractors, respectively. We extract feature maps \(f_{ori}\) and \(f_{les}\) using these two CNNs. Then, \(f_{ori}\) and \(f_{les}\) are concatenated on channel dimension as the input of the attention network.
The attention network consists of a \(3\times 3\) Conv, a ReLU, a dropout, a \(1\times 1\) Conv, and a Sigmoid layer. It produces two weight maps \(W_{ori}\) and \(W_{les}\), which have the same shape as the feature maps \(f_{ori}\) and \(f_{les}\), respectively. Then, we compute the weighted sum f(i, j, c) of the two feature maps as follows:
where \(\circ \) denotes element-wise product. The weights \(W_{ori}\) and \(W_{les}\) are computed as \( W(i,j,c) = \frac{1}{1+e^{-h(i,j,c)}} \), where h(i, j, c) is the last layer output before Sigmoid produced by the attention network. W(i, j, c) reflects the importance of the feature at position (i, j) and channel c. The final output is produced by performing a softmax operation on f(i, j, c) to get the probabilities of all grades.
3 Experiments
In this section, we evaluate Center-Sample detector and AFN on various datasets.
3.1 Evaluating Center-Sample Detector
Dataset and Evaluation Metric. A private dataset was provided by a local hospital, which contains 13k abnormal (more severe than grade of 0) fundus images of size about \(2000\times 2000\). Lesion bounding boxes were annotated by ophthalmologists, including 25 k MA and 34 k HE lesions, with about 26% missing annotated lesions. The common metric for object detection mAP is used as the evaluation metric since it reflects the precision and recall of each lesion.
Implementation Details. In our experiments, we select MA and HE lesions as the detection targets since other types of lesions are clear even in compressed images (\(512\times 512\)). During training, we train the model with cropped patches (\(300\times 300\)) which include annotated lesions from the original images. Random flips are applied as data augmentation. We use SGD (momentum = 0.9, weight decay = \(10^{-5}\)) as the optimizer and batch size is 16. The learning rate is initialized to \(10^{-3}\) and divided by 10 after 50k iterations. When training the Center-Sample detector, we first use center loss and detection loss as multi-task loss for pre-training. Then the Center-Sample mechanism is included after 10k training steps. \(t_l\) and \(t_u\) are set to 1st and 5th percentile among all deep features in one batch.
Results and Analysis. We evaluate the effects of the Center-Sample components by adding them to the detection model one by one. Table 1 shows that the base detection network (BaseNet), which is similar to SSD, gives mAP = 41.7%. After using Center Loss as one part of multi-task loss, it raises to 42.2%. The Center-Sample strategy further adds 1.4% to it, with the final mAP = 43.6%. Note that common detectors like SSD lack mechanisms to address the missing annotation issue. The results show the robustness of our proposed method. Figure 3 visualizes some regions where deep features are close to lesion centers.
3.2 Evaluating the Attention Fusion Network
Datasets and Evaluation Metric. The private dataset used (which is different from the one for evaluating Center-Sample above) contains 40k fundus images, with 31k/3k/4k/1.1k/1k images for DR grades 0 to 4 respectively, rated by ophthalmologists. The EyePACS dataset gives 35k/11k/43k images for train/val/test sets, respectively. The Messidor dataset has 1.2 k retinal images with different criteria for DR grades 0 to 3. For the EyePACS dataset and the private dataset, we adopt the quadratic weighted kappa score which can effectively reflect the performance of the model on an unbalanced dataset. For the Messidor dataset, we refer to the experimental methods [11] and conduct tasks of referable v.s. non-referable and normal v.s. abnormal, with AUC as the metric.
Implementation Details. We use two ResNet-18 [6] as the feature extractors for both inputs. The preprocessing includes cropping the images and resizing them to \(224\times 224\). Random rotations/crops/flips are used as data augmentation. AFN is trained with the SGD algorithm. All models are trained for 300k iterations with the initial learning rate=\(10^{-5}\) and divided by 10 at iterations 120k and 200k. Weight decay and momentum are set to 0.1 and 0.9.
Results on the Private Dataset. We evaluate AFN and several models on the private dataset as shown in Table 2. Baseline only employs scaled original images as input to ResNet-18 for training. We re-implement the feature fusion method in [13], called Two-stage. Another fusion method that concatenates lesion maps and scaled images on channel dimension (called Concated) is compared, since both these inputs equally contribute to identifying DR with this method. Our approach outperforms the other methods considerably. Note that the Two-stage method performs not as well as in the original paper [13] on our dataset, possibly for the following reasons. (a) The Two-stage method cannot identify grade 4 well, because MA and HE lesions might be noisy information for grade 4. (b) There are some unannotated lesions in our dataset. We visualize F1 scores of identifying PDR (grade 4) in Fig. 4, which shows AFN has similar ability as Baseline to determine PDR, and other models perform better than Baseline as a whole but worse in PDR identification. This shows the lesion maps of MA and HE are useless noisy information for PDR and our AFN can reduce the impact.
Results on EyePACS and Messidor. We use the Center-Sample detector trained on the private datasets to produce EyePACS and Messidor’s lesion maps. Table 3 shows that AFN obtains kappa scores of 0.857 and 0.849 on the val/test sets, respectively. Since the size of Messidor is quite small for training CNNs from scratch, we fine-tune AFN using weights pre-trained on EyePACS. Table 4 shows the results of proposed approach compared with previous studies. To our best knowledge, we achieve state-of-the-art results on both public datasets.
4 Conclusions
In this paper, we proposed a new framework unifying lesion detection and DR grade identification. With the Center-Sample detector, we can use low quality annotated data to train an effective model, and employ center loss to make the model more discriminative and robust. Further, using a new information fusion method based on attention mechanism, we achieve better DR identification. Experiments showed that our approach outperforms state-of-the-art methods.
References
Chen, L.C., Yang, Y., Wang, J., Xu, W., Yuille, A.L.: Attention to scale: Scale-aware semantic image segmentation. In: CVPR, pp. 3640–3649 (2016)
Dai, L., et al.: Retinal microaneurysm detection using clinical report guided multi-sieving CNN. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10435, pp. 525–532. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66179-7_60
Gargeya, R., Leng, T.: Automated identification of diabetic retinopathy using deep learning. Ophthalmology 124(7), 962–969 (2017)
van Grinsven, M.J.J.P., van Ginneken, B., Hoyng, C.B., Theelen, T., Sanchez, C.I.: Fast convolutional neural network training using selective data sampling: application to hemorrhage detection in color fundus images. IEEE Trans. Med. Imaging 35(5), 1273–1284 (2016)
Gulshan, V., et al.: Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 316(22), 2402 (2016)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: CVPR, pp. 770–778 (2015)
Kumar, N., Rajwade, A.V., Chandran, S., Awate, S.P.: Kernel generalized-gaussian mixture model for robust abnormality detection. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10435, pp. 21–29. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66179-7_3
Liu, W., et al.: SSD: single shot multibox detector. In: Leibe, B., Matas, J., Sebe, N., Welling, M. (eds.) ECCV 2016. LNCS, vol. 9905, pp. 21–37. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46448-0_2
Seoud, L., Hurtut, T., Chelbi, J., Cheriet, F., Langlois, J.M.P.: Red lesion detection using dynamic shape features for diabetic retinopathy screening. IEEE Trans. Med. Imaging 35(4), 1116–1126 (2015)
Sánchez, C.I., Niemeijer, M., Dumitrescu, A.V., Suttorpschulten, M.S., Abràmoff, M.D., Van, G.B.: Evaluation of a computer-aided diagnosis system for diabetic retinopathy screening on public data. IOVS 52(7), 4866 (2011)
Wang, Z., Yin, Y., Shi, J., Fang, W., Li, H., Wang, X.: Zoom-in-net: deep mining lesions for diabetic retinopathy detection. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10435, pp. 267–275. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66179-7_31
Wen, Y., Zhang, K., Li, Z., Qiao, Y.: A discriminative feature learning approach for deep face recognition. In: Leibe, B., Matas, J., Sebe, N., Welling, M. (eds.) ECCV 2016. LNCS, vol. 9911, pp. 499–515. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46478-7_31
Yang, Y., Li, T., Li, W., Wu, H., Fan, W., Zhang, W.: Lesion detection and grading of diabetic retinopathy via two-stages deep convolutional neural networks. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10435, pp. 533–540. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66179-7_61
Acknowledgement
D.Z. Chen’s research was supported in part by NSF Grant CCF-1617735. The authors would like to thank the RealDoctor AI Research Center.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Lin, Z. et al. (2018). A Framework for Identifying Diabetic Retinopathy Based on Anti-noise Detection and Attention-Based Fusion. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11071. Springer, Cham. https://doi.org/10.1007/978-3-030-00934-2_9
Download citation
DOI: https://doi.org/10.1007/978-3-030-00934-2_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00933-5
Online ISBN: 978-3-030-00934-2
eBook Packages: Computer ScienceComputer Science (R0)