Abstract
There exist several dissemination repositories, computation platforms, and online tools that might be used to implement Reproducible Research. In this paper, we discuss the strengths and weaknesses, or better, the adequacy of each of them for this purpose. Specifically, we present aspects such as the freely availability of contents for the scientific community, the languages which are accepted, or how the platform solves the problem of dependency to specific library versions. We discuss if articles and codes are peer-reviewed or if they are simply spread through a dissemination platform, and if changes are allowed after publication. The most popular platforms and tools are presented with the perspective to highlight new ways for scientific communication.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
Reproducibility and replicability are common and usual criteria in many fields as biology, economy, psychology or medicine. Although paramount, meeting these criteria is a goal difficult to achieve. In Computer Science, it is easier since, most of the time, it involves applying the same data to the same algorithm. Ten years ago, Donoho et al. mentioned the so-called credibility crisis and suggested to not only publish computational results but also the complete source code and data [1]. More recently, this crisis was also mentioned in a research survey where more than 70% of researchers affirm that they fail to reproduce the work of another researcher [2]. Moreover, more than half affirm they fail reproduce their own work. This survey was realized in different domains such as biology, physics and, chemistry.
Before the awareness of this problem, the benefit of Reproducible Research (RR) for authors and computational scientists was illustrated with concrete example from Wavelet Matlab package. According to Buckheit et al. [3], the package was designed to help implement the reproducible research following the guidelines of Claerbout [4], who defines the scholarship in computational science not as the scientific publication but as “the complete software development environment and the complete set of instructions which generated the figures”. As shown in the following, RR presents an increasing importance for platforms, in the J. Buckheit et al. spirit, with the perspective to define a new way of diffusing scholarship contents.
Several definitions of a reproducible result were proposed. In the following, we consider the definitions by Krijthe and Loog [5] and by Rougier and Hinsen [6]. These authors distinguish two main notions, the reproducibility and replicability. The reproducibility is defined as the success of a same individual in re-generating the same results with the same inputs and following the same protocols. The replicability reflects the possibility to obtain identical results in a different context without taking the same implementation and protocol of the first authors. It can be considered as valuable since it offers the proof that the proposed scholarship is not only specific to particular conditions but can also be extended to other kinds of situations.
Mainly motivated by reproducible and replicable aspects, new platforms are appearing to help researchers to execute, reproduce, experiment, and compare or diffuse research codes and software, often through an online website. In parallel, other initiatives are growing with introduction of new journals focused on original content as softwares associated to research publications, replication experiences, or specialized in the image processing domain. In the following of this paper, we first present an overview of these platforms (Sect. 2) before comparing them through several criteria (Sect. 3). In addition, we will focus on new initiatives like the Graphics Replicability Stamp Initiative that was proposed in conjunction to more classic conferences in order to augment publication with reproduction guarantee (Sect. 4).
2 Overview of Reproducible Research Platforms
In this section we present different types of platforms that could be used to implement RR. We refer to them as:
-
Online execution platforms (Sect. 2.1): an online platform offering an execution service infrastructure.
-
Dissemination platforms (Sect. 2.2): their goal is to spread articles, source code, data, and make them public, without necessarily being peer-reviewed.
-
Peer-reviewed journals (Sect. 2.3): similar to the dissemination platforms, but with an Editorial Board that requests opinion of external experts to decide article acceptation.
2.1 Online Execution Platforms
Some very well-known platforms are closely related to RR in there usages, whether they are domain-specific or more generic.
Galaxy. In the case of Biology, the Galaxy project [7,8,9] started in 2005 as a platform for genomics research making available tools which can be used by non-expert users too. Galaxy defines a workflow as a reusable template containing different algorithms applied to the input data. In order to achieve reproducibility, the system stores the input dataset, the parameters, the tools and algorithms applied to the data within the chain, and the output dataset. Thus, performing the same workflow with the same data ensures that the same results are obtained, given that the version of all the elements remains the same. The platform allows its users to upload their own data and to adjust the parameters before executing an algorithm. Galaxy is made of four main elements: (i) the main public Galaxy server featuring tool sets and data for genomics analysis, (ii) open source software and API allowing users to install their own Galaxy server, (iii) a repository for developers and administrators, and (iv) the whole community contributing to the development.

extracted from https://github.com/galaxyproject/galaxy.
The GitHub activity (in number of commits) of the main Galaxy project,
As shown in Fig. 1, the activity of the Galaxy platform is increasing from its beginning and up to now. The Galaxy community has a hubFootnote 1 that includes yearly conferences (Galaxy Community Conference). This community is also composed of several groups associated to the Intergalactic Utilities Commission, a Galaxy training network and an open source package management system. Some regional communities are visible from several countries (Arabic, Austria, France, Japan, UK).
IPython. Generic tools for RR include the IPython tool and its notebooks [10]. This mature tool created in 2001 allows to create reproducible articles, not only by editing text in the notebook, but also by allowing code execution and creating figures in situ. This approach follows closely the definition of a reproducible scientific publication given by Claerbout. Even though the high activity of the platform was during 2011–2015 (from the GitHub remote source), this platform is always active and continues to share the main kernel of the Jupyter platforms described below.
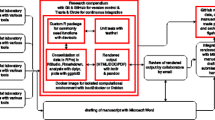
Jupyter. In 2014 the Jupyter project [11] was started as a spin-off of IPython in order to separate the Python language used in IPython from all the other functionalities needed to run the notebooks (for example, the notebook format, the web framework, or the message protocols) [12]. IPython turns then into just another computation kernel for Jupyter, which nowadays supports more than 40 languages that can be used as kernelsFootnote 2. The Jupyter notebook also provides new ways to generate interactive webpages from the nbinteract viewer [13]. The Fig. 2 illustrates an example of Jupyter notebook.
Jupyter: The Binder Service. Directly related to the Jupyter platform, this service offers the possibility to transform any repository containing a Jupyter notebook source into an online notebook environment. Therefore, user experiments can be reproduced and executed from a simple git repository without installing any Jupyter server. The Fig. 3 illustrates the three main steps to reproduce the results of a demonstration code from a git repository. Note that the service is free of charge and no registration is needed to build or run the resulting notebook.
Runmycode.online. In the same line than the previous Binder service, this tool is able to execute a source code hosted on an external repository [14]. The website proposes a navigator extension that allows to set the program parameters and start running the code. It can be run online from various code repositories as GitHub, GitLab, or BitBucket, and accepts several languages such as C/C++, Java, Nodejs, python. PHP, Ruby, and others. The service is free to use.
Code Ocean. Started in February 2017, this platform [15] is a recent initiative from the IEEE with the aim to attach online demonstrations to the published articles. It defines itself as a cloud-based computational reproducibility platformFootnote 3. The platform is only designed to run code and not to really publish article even if a DOI is assigned to each source code. Numerous languages are accepted (Python, R, Julia, Matlab, Octave, C++, Fortran, Perl, Java, Sata, Lua, Octave). There are fees to pay according to the computational load of the servers running the algorithms. A free plan exists, but limited to two concurrent users and 1h/month of computing time.
Note that recently, three Nature journals have run a trial partnership with this platform, allowing authors to demonstrate the reproducibility of their works [16]. Such an initiative could be an interesting answer to the reproducibility crisis mentioned in introduction.
Development environment of the Code Ocean platform,
The Fig. 4 illustrates the Code Ocean platform that can be executed online. The main view of the platform is composed of sub-views showing the inputs and the results. Note that the source code can also be edited by the user, in a private environment.
The DAE Platform. In the field of document image analysis, the Document Analysis and Exploitation platform (DAE) was designed to share and distribute document image with algorithms. Created in 2012, the DAE platform contains tools to exploit document annotations and to perform benchmark [17]. Similarly to the previous platforms, DAE allows the users to upload their own data and to combine several algorithms. The user can tune the parameters of each of the algorithms proposed by the platform. The users have control over the data uploaded, the parameters used, and the choice of the platform on which the algorithms are available. The Fig. 5 illustrates an example of the DAE data sets with different scanned documents. The code is executed exclusively online but is free to use and without any computing time limitation (contrary to Code Ocean). The source code of the platform is also open source and available onlineFootnote 4.
Example of database available on the DAE platform, at address http://dae.cse.lehigh.edu/DAE/?q=browse
The IPOL Journal. It is a full peer-reviewed indexed journal where each paper is associated to an online demonstration and an archive of experiments. The set of online demonstrations [18] can be seen as an online execution platform that share some of the characteristics with Jupyter. In particular, IPOL proposes to run algorithms online through a web interface, to obtain an immediate visualization of the results and to write algorithms in several programming languages. Moreover, it is free of any cost for authors and users.
IPOL is however exclusively designed to run the demonstration of already published papers. An original point is the fact that it contains the history of the different executions with original source images and parameters. The online demonstrations are build from a simple demo description language avoiding the author to code the demonstration interface himself.
Regarding the system architecture, IPOL is built as a Service-Oriented Architecture (SOA) made of micro-services. This type of architecture allows to have simple units (called modules in its own terminology, or micro-services) which encapsulate isolated high-level functions. Other examples of service-oriented architectures made of micro-services are Amazon AWS API GatewayFootnote 5, Netflix [19] or Spotify [20].
IPOL and Code Ocean share many features, as a user-friendly demonstration builder, an online code execution, and an advanced visualization. But they also have some differences since IPOL stores all experiments performed by the users in a freely available archive, has demonstrations always free to use, and doesn’t require authentication to execute a demonstration.
2.2 Dissemination Platforms
Other generic tools can be seen as dissemination platforms since their objective is to make source code and data widely available to everyone. This category contains among others:
-
ResearchCompendia [21], that stores data and code in an accessible, traceable, and persistent form.
-
MLOSS [22], dedicated to machine learning.
-
DataHub [23], that allows to create, register, and share generic datasets.
-
RunMyCode [24], that associates code and data to scientific publications.
-
IPOL, that, in addition to propose an online execution environment (see Sect. 2.1), makes available the source code of the algorithm.
The various community code development platforms like GitHub, GitLab, BitBucket can also be considered as dissemination platforms since they contribute to diffuse source codes. However, the evolution of the repositories over time is a key question and no guarantees are given from their owners. Despite this, each user can easily replicate a repository to another platform, thanks to the distributed version-control systems, as git or mercurial, on which they are based.
A more global answer to this problem was given in France, leading by the National Institute in Computer Science and Automation (INRIA), at the origin of a project called Software Heritage. Its aim is to collect repositories from different platforms into one single place with the plan to ensure their durability [25]. This system is linked to HAL Open Archives System [26], an open source paper publication system. The platform de-duplicates any submitted repository from different platforms but do not review any code nor evaluate the execution. An interesting perspective could be to propose a way to obtain a compilation/execution status for each repository. This could be done, for instance, with tools such as Travis [27] or the Docker framework [28].
2.3 Peer-Reviewed Journals
IPOL Journal. The Image Processing OnLine journal was founded in October 2009 at the initiative of Nicolas Limare and Jean-Michel Morel at CMLA (Université Paris-Saclay), with the first paper published in 2010. Motivated by RR and first focused on image processing, it differs from classic journals since all articles are provided with (i) a mathematical description of the method, as detailed as possible, (ii) the source code of the presented algorithms, and (iii) an online demonstration [29]. Each demonstration has an archive that stores the history of all executions performed with data from the users. As in classic journals, it has an Editorial Board and every article is assigned an editor that will put the paper under peer-review (both the article and the source code). A mandatory requirement is that each description (pseudo-codes, formulas) given in the text need to match exactly to the source code implementation.

IPOL is an Open Science journal, with an ISSN and a DOI, that contains more than 140 papers covering various image processing subdomains like image de-noising, stereovision, segmentation, 3D mesh processing, or computer graphics. The journal was extended to video and audio processing, as well to general signal processing, including physiological signals.
The main goal is to establish a complete state of the art in algorithms for general signal processing.

ReScienceJournal. This peer-reviewed journal was created in 2015 at the initiative of Konrad Hinsen and Nicolas Rougier. Their motivation follows the replication main goals, as defined by [30] or [5]. ReScience aims to promote already published work and highlights the reproduction of research results in new or different contexts. The authors of the original work are not allowed to submit their own work, even if they claim to have changed their results, architectures, or frameworks. The submission process takes place from the GitHub platform with a direct non-anonymous interaction through a Pull Request process, used by authors to integrate the new contribution in a shared project. ReScience is an online Open Science journal, with article in PDF form. It has a classic organization with volume number, DOI, and reviews available online on GitHub.
A typical contribution to ReScience is the case of an author who would like reproduce an existing method for which its authors do not propose any implementation. The author can then replicate the method from the original paper and submit his implementation to ReScience, including a description of the main steps to reproduce the results.
The journal contains currently 22 accepted contributions, all of them successful replications. Failing replication experiences can also be potentially accepted for publication, even if so far all published papers concern successful replications. A call for replication is diffused on the GitHub repository, allowing to suggest the replication of published papers [31]. The call is materialized by adding an issue on the repository, from which users can discuss and interact. A more detailed presentation of the journal can be found in a recent article [6].

JOSS Journal. The Journal of Open Source Software was founded by Arfon Smith in May 2016. The motivation to create this new journal comes from the fact that “Current publishing and citation do not acknowledge software as a first-class research output” [32]. To answer this, the journal acknowledges research software and contributes to offer modern computational research results. JOSS is a free Open Science journal and similarly to the ReScience journal, it is hosted on GitHub with a public peer reviewing process that provides direct visibility. As other classic journals it has an ISSN number and the articles a Crossref DOI.
Associated to its source code archive, a JOSS publication contains also a PDF with a short abstract and a short description of the content with a link to the forked repository of the software. The very short description is voluntary since novel research descriptions is not requested (and not allowed) and is not what the journal is focused on. Publications of APIs are neither allowed. The source code needs to be open source and the submitter needs to be main contributor. Another requirement of the journal is to submit a complete and fully functional product.
The journal covers several domains including Astronomy, Bioinformatics, Computational Science, Data Science, General Engineering, among others. It contains currently more than 551 accepted papers. With a basis of 100 papers per year, the editor of the journal estimates the cost around of $6/paper [32]. The current editor strategy is to handle software versions throughout the publication steps. Requesting major new features can be important to maintain the value for the publications itself while minor updates are also welcome to ensure the maintenance of the software.

Insight J Journal. As JOSS, Insight J is an online Open Science journal and covers the domain of medical image processing and visualization [33]. It was created in 2005 by the Insight Software Consortium and contains currently 642 publications with 768 peer open reviews. All the publications do not necessary receive reviews and numerous papers are visible online and do not have any reviews even ten years after publication. A star based rating process is proposed for any user and allows to evaluate the paper, source code and the review quality. A top ranked list of publications is available along with the number of downloads and views.
Contrary to the JOSS journal, Insight J is mainly related to a specific library (ITK) and can be seen as a way to increase the value of contributions to this library. Accepted publications usually propose a new module that can be potentially integrated within the library. It generally contains a scientific description with a detailed description of the implementation proposed, using the ITK framework together. The journal has an ISSN number and all its papers are indexed by Google Scholar, thus allowing to compute a citation score. The number of citations of the most downloaded paper is relatively low compared to other similar journals. For instance, the Insight J paper of Tustison et al. [34] was cited 68 times while, for instance, the popular IPOL paper of Grompone et al. [35] accounts 373 references. However, it appears that the publications in Insight J contribute to promote other associated papers that are published in more general classic journals. For example, an associated paper published in a medical imaging journal [36] obtained 1230 citations.
Notes that two other variants exist with the same form and identical publication process but focused on scientific visualization: the VTK Journal [37] (using VTK library) and the MIDAS journal for visualization and image processing [38].
In the context of RR, these almost new journals offer new complementary alternatives of publication that can break with the usual procedures of scientific communication. The model of the IPOL journal follows a larger content model, including scientific descriptions, online demonstrations and perfect matching between algorithms and source codes. More focused on the replication, ReScience covers a larger domain and publication related to already known results can foster the discussion in the scientific community. Research software can also be published in the JOSS journal whereas contributions on image processing libraries can be proposed to Insight J.
3 Comparison
This section summarizes and confronts the characteristics of the previously presented platforms. It shows their strengths and weaknesses as well as opportunities and eventual threats of used approaches. The following criteria are evaluated:
- (1) :
-
Free to use
- (2) :
-
No mandatory registration
- (3) :
-
Several programming languages allowed
- (4) :
-
Peer-reviewed code and data
- (5) :
-
Easy to use by the non-expert
- (6) :
-
General scope
- (7) :
-
Possibility to upload user data
- (8) :
-
Interaction through a web interface
- (9) :
-
Access to a public and persistent archive of experiments
- (10) :
-
Design of automatic demonstration from textual description or visual tool
- (11) :
-
Allow to modify the source code before execution
Platform | (1) | (2) | (3) | (4) | (5) | (6) | (7) | (8) | (9) | (10) | (11) |
|---|---|---|---|---|---|---|---|---|---|---|---|
Galaxy |
|
|
|
|
|
|
|
|
|
|
|
DAE |
|
|
|
|
|
|
|
|
|
|
|
IPython |
|
|
|
|
|
|
|
|
|
|
|
Jupyter |
|
|
|
|
|
|
|
|
|
|
|
Code Ocean |
|
|
|
|
|
|
|
|
|
|
|
Res. Comp. |
|
|
|
|
|
|
|
|
|
|
|
DataHub |
|
|
|
|
|
|
|
|
|
|
|
RunMyCode |
|
|
|
|
|
|
|
|
|
|
|
IPOL |
|
|
|
|
|
|
|
|
|
|
|
Legend | |
|---|---|
– | Not Applicable |
(A) | It allows users to execute the proposed algorithms, but it is not a dissemination platform and thus it does not accept a list of languages. Instead, it wraps the algorithms and incorporates them |
(B) | It frees the user from using command line tools, but it still requires to know the details of the algorithms to perform genomics analysis |
(C) | A free plan is offered, but limited to 1h/month of computing time, and a single researcher user. With a cost of $20/month, it allows 10h/month and 5 concurrent users |
(D) | A web interface is proposed, but the usability depends on the author who creates its own interface |
(E) | The user interacts through a web interface without interactive demonstration |
(F) | The demonstrations are free to use up to some limits (say, size of the data or computation time), but industrial use of demonstrations and applications requires payment |
(G) | True for demonstrations using a sample learning dataset. To use the platform as a service, the user needs to be connected with a role authorizing this usage |
This comparison of platforms should not be seen as a competition to decide if a platform is better or worse than the others, but as a way to decide which platforms are more adapted to a particular application.
Almost all platforms are free to use (1) with the exception of Code Ocean, which offers a very limited free plan.
The possibility of using a platform without a prior registration (2) greatly helps its diffusion and, in the case of scientific applications, it allows spreading knowledge. Some of platforms are mainly oriented to specialists, as for instance Galaxy (genomics research).
For any of the platforms publishing source code it is important that at least the most popular languages and frameworks are supported (3). In the case of the Galaxy and DAE platforms that do not publish algorithms but use them to offer a service, they solve the problem by wrapping the algorithms. In the case of platforms publishing algorithms like Code Ocean, ResearchCompendia, RunMyCode, and IPOL, they accept the most popular languages, frameworks and libraries.
About the peer-review of the code (4), Galaxy and DAE do it before wrapping and incorporating them into their platform, once the interest and opportunity of making publicly available a particular algorithm is clear. Others like Code Ocean, DataHub, RunMyCode, IPython and Jupyter do not peer-review the code or the data. The code is carefully peer-reviewed only when the platform publishes paper in a journal, like IPOL, or when algorithms are offered as a professional service, like Galaxy or DAE.
In addition to the journal aspect, IPOL can be used as a computational platform. It allows users to create workshops, which are demonstrations without associated paper. The workshops are not publications and, therefore, are not peer reviewed. This can allow to use IPOL as a computational facility in order to, for instance, monetize services. Some platforms such as Galaxy or DAE try to hide the technical details as the direct calls of tools from the command line and they offer instead a web interface (8). Jupyter and Code Ocean also hide the direct interaction with the framework by proposing a web interface acting as a proxy for execution and visualization. IPOL solves the problem by a flexible interface that can be adapted to each application.
Some of the platforms are domain specific (6) like Galaxy or DAE, whereas others are more general. This frees the domain-specific platforms to create different web interfaces depending on the final application but, on the other hand, makes them less flexible.
For online execution platforms willing to reach RR, it is mandatory that they allow users to upload their own data (7). And, indeed, all of them propose this functionality. Another interesting add-on is to have a permanent and public archive of experiments (9). In the case of Galaxy, DAE, Jupyter, Code Ocean, they are no publicly accessible. Only IPOL has a permanent, public and open archive of experiments.
For the platforms which offer demonstrations, an interesting feature is to modify the source code of the method before running the demo (11). This is only found in Code Ocean. Both Code Ocean and IPOL allow to create new demonstrations (10) just with a simple textual description (IPOL) or a visual tool (Code Ocean).
4 Augmented Publications
The classic way of communicating scientific publications is more and more augmented with new emerging initiatives based on the reproducibility. In the domain of Geometry Processing, the Graphics Replicability Stamp Initiative (GRSI) was created in order to certify the reproduction of both the results and figures contained in the published paper [39]. This initiative was created at the thirteenth Symposium on Geometry Processing where it was proposed to authors of accepted papers to apply to this stamp. Note that it was initially known as the Reproducibility Stamp and the new “Replicability” term differs from the definition used in this paper and is in fact related to the reproducibility. The initiative was continued and extended through collaborations with journals ACM TOG, IEEE TVCG, Elsevier CAGD and Elsevier C&G. Currently 32 contributions received the stamp after 3 years of activity. The contributions are all hosted on the GitHub repository.
More related to the Pattern Recognition domain, a similar initiative was proposed with the first workshop on Reproducible Research in Pattern Recognition (RRPR 2016) [40]. This satellite workshop was proposed along the ICPR conference and a reproducible label was given to authors of the main conference after review. The aim of the label is to ensure reproducibility of the paper results, i.e. figures and tables. The labeled contributions are also hosted on GitHub from a fork of the authors repository.
5 Conclusion
This paper presented an overview of the main structures contributing to facilitate reproducible works. Starting from the RR platforms that change potentially the way the information is spread, we explored four main journals that achieve to make original contributions that go beyond classic publications. We also described recent initiatives allowing to extend or augment classic publication procedures, in particular by showing reproducible results in the form of online algorithm executions. Reproducible Research is absolutely needed to avoid fraud, to establish the state of the art in all involved disciplines, and to definitively ensure reliable scientific practices. All the presented platforms make a valuable contribution in this direction and, with all other mentioned initiatives, will certainly help to disseminate good science and reliable knowledge.
References
Donoho, D.L., Maleki, A., Rahman, I.U., Shahram, M., Stodden, V.: Reproducible research in computational harmonic analysis. Comput. Sci. Eng. 11, 8–18 (2009)
Baker, M.: 1,500 scientists lift the lid on reproducibility. Nature 533, 452 (2016). https://doi.org/10.1038/533452a
Buckheit, J.B., Donoho, D.L.: WaveLab and reproducible research. In: Antoniadis, A., Oppenheim, G. (eds.) Wavelets and Statistics. Lecture Notes in Statistics, vol. 103, pp. 55–81. Springer, New York (1995). https://doi.org/10.1007/978-1-4612-2544-7_5
Claerbout, J.F., Karrenbach, M.: Electronic documents give reproducible research a new meaning, pp. 601–604 (2005)
Krijthe, J.H., Loog, M.: Reproducible pattern recognition research: the case of optimistic SSL. In: Kerautret, B., Colom, M., Monasse, P. (eds.) RRPR 2016. LNCS, vol. 10214, pp. 48–59. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-56414-2_4
Rougier, N.P., Hinsen, K.: ReScience C: a journal for reproducible replications in computational science. In: Kerautret, B., et al. (eds.) RRPR 2018. LNCS, vol. 11455, pp. 150–156. Springer, Cham (2019)
Giardine, B., et al.: Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15, 1451–1455 (2005)
Afgan, E., et al.: The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 44, W3–W10 (2016)
Goecks, J., Nekrutenko, A., Taylor, J.: Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11, 1 (2010)
Pérez, F., Granger, B.E.: IPython: a system for interactive scientific computing. Comput. Sci. Eng. 9, 21–29 (2007)
et al., P.: Jupyter Project (2018). http://jupyter.org
Kluyver, T., et al.: Jupyter Notebooks? A publishing format for reproducible computational workflows. In: Loizides, F., Scmidt, B. (eds.) Positioning and Power in Academic Publishing: Players, Agents and Agendas, pp. 87–90. IOS Press, Amsterdam (2016)
Lau, S., Hug, J.: nbinteract: generate interactive web pages from Jupyter notebooks. Master’s thesis, EECS Department, University of California, Berkeley (2018)
Gupta, S.: RunMyCode (2017). https://runmycode.online. Accessed Apr 2019
Code Ocean (2019). https://codeocean.com/. Accessed May 2019
Staniland, M.: Nature research journals trial new tools to enhance code peer review and publication. Nature.com Blogs (2018)
Lamiroy, B., Lopresti, D.P.: The DAE platform: a framework for reproducible research in document image analysis. In: Kerautret, B., Colom, M., Monasse, P. (eds.) RRPR 2016. LNCS, vol. 10214, pp. 17–29. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-56414-2_2
Arévalo, M., Escobar, C., Monasse, P., Monzón, N., Colom, M.: The IPOL Demo system: a scalable architecture of microservices for reproducible research. In: Kerautret, B., Colom, M., Monasse, P. (eds.) RRPR 2016. LNCS, vol. 10214, pp. 3–16. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-56414-2_1
Izrailevky, Y.: Completing the netflix cloud migration (2016). https://media.netflix.com/en/company-blog/completing-the-netflix-cloud-migration. Accessed Apr 2019
Goldsmith, K.: Microservices at spotify (2016). https://es.slideshare.net/kevingoldsmith/microservices-at-spotify. Accessed Apr 2019
Research Compendia (2019). http://researchcompendia.science/. Accessed Apr 2019
MLOSS (2019). http://mloss.org. Accessed Apr 2019
Datahub (2019). https://datahub.io/. Accessed Apr 2019
RunMyCode (2019). http://www.runmycode.org/. Accessed Apr 2019
Cosmo, R.D., Zacchiroli, S.: Software heritage: why and how to preserve software source code. In: iPRES 2017: 14th International Conference on Digital Preservation, Kyoto, Japan (2017). https://www.softwareheritage.org
Baruch, P.: Open Access Developments in France: the HAL Open Archives System. Learned Publishing 20, 267–282 (2007). See also; voir aussi: P. Baruch, La diffusion libre du savoir Accès libre et Archives ouvertes. http://archivesic.ccsd.cnrs.fr/sic_00169330/fr/
Travis (2019). https://travis-ci.com. Accessed May 2019
Docker (2019). https://www.docker.com/. Accessed May 2019
Colom, M., Kerautret, B., Limare, N., Monasse, P., Morel, J.M.: IPOL: a new journal for fully reproducible research; analysis of four years development. In: 2015 7th International Conference on New Technologies, Mobility and Security (NTMS), pp. 1–5. IEEE (2015)
Patil, P., Peng, R.D., Leek, J.: A statistical definition for reproducibility and replicability. bioRxiv (2016)
GitHub Call for replication repository (2016). https://github.com/ReScience/call-for-replication
Smith, A.M., et al.: Journal of open source software (JOSS): design and first-year review. PeerJ Comput. Sci. 4, e147 (2017)
Insight J: Insight journal (2019). ISNN 2327–770X. http://insight-journal.org
Tustison, N., Gee, J.: N4ITK: Nick’s N3 ITK implementation for MRI bias field correction. Insight J. 9 (2009)
Von Gioi, R.G., Jakubowicz, J., Morel, J.M., Randall, G.: LSD: a fast line segment detector with a false detection control. IEEE Trans. Pattern Anal. Mach. Intell. 32, 722–732 (2010)
Tustison, N.J., et al.: N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging 29, 1310 (2010)
The VTK Journal (2019). ISSN 2328–3459. https://www.vtkjournal.org
The MIDAS Journal (2019). ISSN 2182–95432. https://midasjournal.org
Panozzo, D.: Graphics replicability stamp initiative (2016). http://www.replicabilitystamp.org. Accessed Apr 2019
Kerautret, B., Colom, M., Monasse, P. (eds.): RRPR 2016. LNCS, vol. 10214. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-56414-2
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Colom, M., Kerautret, B., Krähenbühl, A. (2019). An Overview of Platforms for Reproducible Research and Augmented Publications. In: Kerautret, B., Colom, M., Lopresti, D., Monasse, P., Talbot, H. (eds) Reproducible Research in Pattern Recognition. RRPR 2018. Lecture Notes in Computer Science(), vol 11455. Springer, Cham. https://doi.org/10.1007/978-3-030-23987-9_2
Download citation
DOI: https://doi.org/10.1007/978-3-030-23987-9_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-23986-2
Online ISBN: 978-3-030-23987-9
eBook Packages: Computer ScienceComputer Science (R0)