Abstract
Tuberculosis (TB) is one the leading killers in the world, and its early detection at scale is a challenge that remains. Computer Aided Detection of Tuberculosis is an important possibility for the world due to the mismatch in the incidences of this disease with the number of trained human readers for its identification. In this paper, we propose novel features for the detection of one of the symptoms observed in cases of TB, Pleural Effusion (PE). We begin by segmenting the lung regions, followed by creation of a novel feature set. We achieve an ROC of 0.961 on discriminating PE against Chest X-Rays (CXRs) without incidences of TB. To validate that our system discriminates against PE, we achieve an ROC of 0.864 against CXRs showing incidences of TB but a lack of PE. These features are then tested on two publicly available datasets (One collected from the United States, and the other from China). Due to the lack of other work for detection of PE on these datasets, a direct comparison is unfortunately not possible. However, the results obtained surpass those of work on PE detection on other private datasets.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Tuberculosis (TB) is one of the world’s leading killers, with a mortality rate of 1.2 million people in 2010 [1]. While being curable, identifying it in its early stages on scale is a challenge that remains. While the gold-standard tests for TB are slow, Chest X-Rays (CXRs) form the first phase of test for TB, and work as an inexpensive preliminary screening method [2]. However, the interpretation of the CXR depends on the skill of the reader, and is subject to human error and biases. As a result, computer aided detection of tuberculosis through analysis of CXRs seems like a promising idea. This will result in a more easily accessible preliminary screening, and reduce the possibility of errors.
Over the years, much work has been done towards detection of abnormalities in CXRs, but only a few have been geared towards specifically detecting TB. Kuo et al. [3] propose a method for detection of abnormalities (including TB) in CXRs, through the use of novel roughness and symmetry measures for parts of the lung. For specifically discriminating against TB, Jaeger et al. [4] identify TB through a combination of shape and texture descriptors along with the features based on the Lucene Image Retrieval Library. Ginneken et al. [5] begin off by subdividing the lung into smaller regions followed by texture analysis. More information on TB screening systems can be found in the survey by Jaeger et al. [6].
In this paper, we use a different approach to work towards identifying TB in CXRs. We plan to look at the symptoms radiologists tend to look at while reading CXRs, and build classifiers for them individually. Here, we look at the challenge of identifying pleural effusion (PE). While PE by no means is a phenomenon exclusive to TB, it sits in line with our larger aim of building a system to automatically detect TB through analysis of CXRs. This approach would allow a more thorough diagnosis where not only will the overall system assign a score of TB, it will also diagnose the CXR with the correct symptoms.
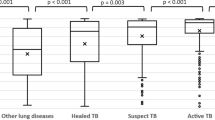
PE is characterized by the buildup of pleural fluid in the lung regions. This fluid tends to accumulate near the bottom of the lungs and the chest cavity [7]. Refer to Fig. 1 to view the manifestations of PE at different severity levels. Limited research has been done on automated detection of PE. Avni et al. [8] proposed a bag of visual words approach to discriminate between various pathologies in CXRs, including a very small dataset of PE. Maduskar et al. [9] work towards identifying the costophrenic point with great accuracy followed by building features around that point. In this paper, we capture the signature of PE using novel features we define after consulting with radiologists on their methods of reading CXRs.
The rest of the paper is structured as follows. In Sect. 2 we explain the process of segmentation of lungs, creation of the features, and details of the classifier. In Sect. 3, the details of the dataset being used are mentioned. Section 4 talks about the results of the experiments. Section 5 deals with the conclusion of the paper.
2 Method
This section talks about our implemented methods for lung segmentation, feature computation, and classification. The system uses a graph cut method to segment the lung, computes the novel features proposed in this work, and then feeds them to a random forest classifier which identifies thresholds around which to classify CXRs.
2.1 Lung Segmentation
The first task of PE detection method is to segment the region of lungs from the CXRs. Since the manifestations of PE occur near the lower boundaries of the lung, it places high requirements on the quality of the segmentation. We initially tried methods based on multilevel thresholding and region growing. Post this, we used a graph cut based segmentation method mentioned in [10]. The method consists of three main steps. It begins with content based image retrieval using a training set along with its defined masks. Post this, the initial patient specific anatomical model is created using SIFT-flow for deformable registration of training masks for the patient CXR. In the final step, a graph cuts optimization procedure with a custom energy function is used. The code made available by Candemir et al. [10] was used for the segmentation process, and hence further details may be inferred from their work. The results of the lung segmentation were good enough to allow us to proceed to the feature designing stage. Figure 2 shows an example of a CXR before and after segmentation.
2.2 Features
The second stage involved crafting features which can discriminate PE. Through consultation with various radiologists, we arrived at a set of visual features they would look for and worked on translating them into their mathematical counterparts. The one valuable insight we gained was that radiologists tend to compare the left and right lung to identify anomalies. The effect of this insight influenced our design of the features.
Another important point to note is that PE is the accumulation of pleural fluid in the lungs, which accumulates in the bottom portion of the lung. This manifests itself as a white region in the CXR, which if large enough, is ignored by the segmentation process as not a part of the lung. We exploit this ‘shortcoming’ to design some of our features.
Height Difference. If the accumulation of the fluid is severe enough, the size of the affected lung is reduced due to its fluid filled part being ignored by the segmentation process. This can be observed in Fig. 3. Hence we take the difference in the heights of the left and right lungs as one of the features. The height of a lung is defined as the distance between the top and bottom pixel of the lung after segmentation. Let \(H_l\) and \(H_r\) be defined as the heights of the left and right lung respectively. Then the feature \(H_{diff}\) is defined as:
We normalise by the maximum of the two heights to account for the fact that people have different sized lungs, and we don’t want the difference to be magnified or reduced solely on the basis of lung size (or even the size of the CXR).
A possible issue which might arise is that the patient might have manifestations of PE in both of his lungs, which results in no height difference. However, such a case is extremely unlikely as a height reduction is observed in severe cases of PE, and such a manifestation in both lungs would be rare. Even if it did occur, such an occurrence of PE would also show up in the other features defined below.
Lung Bottom Curvature. The fluid accumulated at the bottom of the lung settles due to gravity with a flat horizontal surface. This manifests itself as a nearly horizontal white line in the CXR. This affects the segmentation of the lungs, and results in a less curved cut at the bottom after segmentation than normal.
Let \(B_r\) denote the row indexes of the bottom most pixels of columns containing the right lung. It should be noticed that while most of the bottommost pixels of each column will actually be the bottom of the lung, some of them might be the sides of the lung owing to the curvature of the lung. As a result, we disregard 10% of the values on each end of the \(B_r\) array. This figure of 10% was experimentally determined and it may need tuning for other datasets.
Let \(B_l\) denote the row indexes of the bottom most pixels of half the columns on the left side (i.e. only use the left half of the left lung to find row indexes) which contain the left lung. The reason we disregard the right half of the left lung is that a great variation is observed in the row indexes on this end during segmentation due to the presence of the heart, which results in a wide variance of the row indexes which may possibly hide the lack of variance which might be there due to pleural effusion. We also disregard 10% of the values on the other end for reasons similar to case of the right lung.
The feature \(V_r\) is defined as:
where the numerator is the variance of the elements in \(B_r\) and the denominator is the number of elements in \(B_r\). The normalisation by the length of \(B_r\) prevents the variance from being increased or decreased due to individual variation in lung sizes. \(V_l\) is similarly defined for the left lung. These two features are fed to the classifier.
Lower Lung Intensity Variation. In non-severe cases of pleural effusion, a marginal deposition of the fluid in the bottom corners of the lungs is observed, which manifests itself into whiteness in the CXRs. A horizontal scan of the pixel values for each lung starting from the bottom is done, and the mean intensity of the first 7% pixels is calculated. The figure of 7% was arrived at by the desire to capture a large portion of the bottom part of lungs, but at the same time not include too much of the higher portions. The exact number was experimentally determined, and might needed to be tuned for other datasets. The mean intensity of the rest of the pixels is also calculated, and the ratio of the former with respect to the latter is taken. Let \(I_l\) and \(I_r\) denote these values for the right and left lung respectively.
\(I_m\) is one of the features fed to the classifier and is defined as:
\(I_d\) is the other feature used and is defined as:
These two features don’t need to be normalised unlike other features because the normalisation is inherent in the calculation of the \(I_l\) and \(I_r\) itself.
2.3 Classification
After constructing the features, we ended up with a 5 dimensional feature vector for each CXR. These feature vectors were then fed into a random forest classifier [11]. The standard MATLAB implementation of random forest was used for the purpose of the experiments [12].
A random forest classifier is an ensemble learning method which works by constructing multiple decision trees. Each of the decision trees is trained on a different subset on the whole training data, a method known as bootstrap aggregating. The output of the classifier system is taken as the mode of the output of the individual trees which constitute the random forest.
There are a few advantages of random forests which led to this being our classifier of choice. It can learn complex relationships between variables and requires minimal tuning, as opposed to some other classifiers such as SVM. It doesn’t require too much data for learning, unlike neural networks. Additionally, the ensemble of trees help avoid the issue of over-fitting to the data.
While we currently look to classify CXRs on whether they have PE or not, it is also possible in future work to recognise the area where PE has occurred. Since PE mainly occurs near the bottom of the lungs, we need to identify which lungs are affected by PE. This can be done by designing similar features to those above but which aren’t agnostic to the right and left lung. However, this would require a greater amount of data for learning due to increased dimensionality of the features, and hence not explored here.
3 Data
The datasets used in this study were two publicly available CXR dataset provided by the US National Library of Medicine [13]. The first dataset (Montgomery Dataset) contained 138 CXRs with 58 of them showing instances of TB. Each CXR had a dimension of 4020 \(\,\times \,\) 4892 pixels. The second dataset (China Dataset) contained 662 CXRs with 336 of them showing instances of TB. The CXRs in this set had varying dimensions, but were roughly in the range of 3000 \(\,\times \,\) 3000 pixels.
While the sets came with information regarding the presence of TB or not in the CXRs, they did not specifically talk about PE. For the ground truth regarding PE, the dataset was curated by an eminent radiologist. The data was partitioned into three sets. The first set consisted of CXRs which showed instances of TB along with manifestations of PE. The second set consisted of normal CXRs which showed no instances of TB. The third set consisted of CXRs which showed instances of TB but not any manifestations of PE. The number of CXRs showing instances of PE were limited to 63 CXRs, which were a mix from the China and Montgomery dataset. The other two sets were also kept the same size to this to prevent unbalanced classes, which could affect the training of the classifier.
Owing to the less amount of data available, it would not be a good idea to partition the entire data into separate testing and training sets. However, at the same time, it is not feasible to have any common data in the testing and training sets. So, leave-one-out-cross-validation was the preferred method of choice to avoid this issue during classification. It has the advantage of effectively increasing the amount of data for testing purposes, preventing overfitting, and avoiding the excessive computation issue faced in leave-out-p-cross-validation.
4 Results
The performance of the proposed PE detection system was analysed in terms of area (AUC) under the receiver operating characteristics (ROC) curve. As mentioned before, random forests were the classifiers used in the evaluation of performance. The classification accuracy on the PE set is measured against two sets, normal CXRs and those CXRs with manifestations of TB but not PE.
The results of classifying normal CXRs v/s those with PE can be seen in Fig. 4a. We report and AUC of 0.961. Since it is important to not miss TB, we would err on the side of caution and aim for higher sensitivity. The optimal operating point is shown in the ROC curve with a red circle, suggesting we operate at 100% sensitivity and 80.95% specificity. Sensitivity, also known as recall, is the true positive rate of the classifier. Specificity is the true negative rate. This corresponds to a precision (positive predictive value) of 84%.
To ensure that the classifier was learning pleural effusion specific features and not just tuberculosis, we also tested the classifier on the dataset which contained CXRs with manifestations of TB without instances of pleural effusion and the dataset which contained CXRs with manifestations of TB resulting in symptoms of pleural effusion. Even in this case, the classifier achieved an AUC of 0.864 and the ROC characteristics can be seen in Fig. 4b. The optimum point for operation here is at a sensitivity (recall) of 80.95% with a specificity of 77.8%. This corresponds to a precision of 78.4%. If we were to look for 100% sensitivity so as not to miss any manifestation of PE, our precision would fall to around 65%.
The latest work against which we evaluate our results is of Maduskar et al. [9]. They built a system to identify PE by creating features around the costrophrenic point. They evaluate the performance of the system independently for the left and right lungs. However, due to the dataset constraints on our side, this is not a feasible option for us. Their system returns an AUC of 0.84 and 0.90 for PE detection in the left and right lung respectively. This is lower than our AUC reported above. They also evaluate the performance of their system against cases of PE they classify as severe, a level above obvious. They report AUC of 0.88 and 0.94 for the left and right lungs respectively. This is again lower than the AUC of our system.
5 Conclusion
In this study, we have presented a system to detect PE in CXRs. We began by discussing with radiologists their methodology of reading CXRs, and looked to transform this into mathematical formulations. Before extracting features based on these formulations, the left and right lung were separated from the CXR based on a three step segmentation method. Post extracting the features, they were fed into a random forest classifier to arrive on decision boundaries to decide on whether a particular CXR shows instances of PE.
The AUCs of 0.961 and 0.864 are quite encouraging. They are better than the results we have seen in other papers. However, a direct comparison is not possible due to them being based on different datasets, and results being dependent on the datasets. In further work, one can look to build systems to identify other symptoms which manifest in the presence of TB, and then finally look to combine them into one main system.
References
World Health Organization: Global tuberculosis report (2012)
Toman, K., Frieden, T., World Health Organization: Toman’s Tuberculosis: Case Detection, Treatment, and Monitoring: Questions and Answers, 2nd edn. World Health Organization, Geneva (2004)
Kao, E.-F., Kuo, Y.-T., Hsu, J.-S., Chou, M.-C., Liu, G.-C.: Zone-based analysis for automated detection of abnormalities in chest radiographs. Med. Phys. 8(7), 4241–4250 (2011)
Jaeger, S., Karargyris, A., Candemir, S., Folio, L., Siegelman, J., Callaghan, F., Xue, Z., Palaniappan, K., Singh, R.K., Antani, S., Thoma, G., Wang, Y.-X., Pu-Xuan, L., McDonald, C.J.: Automatic tuberculosis screening using chest radiographs. IEEE Trans. Med. Imaging 33(2), 233–245 (2014)
van Ginneken, B., Katsuragawa, S., ter Haar Romeny, B.M., Doi, K., Viergever, M.: Automatic detection of abnormalities in chest radiographs using local texture analysis. IEEE Trans. Med. Imag. 21(2), 139–149 (2002)
Jaeger, S., Karargyris, A., Candemir, S., Siegelman, J., Folio, L., Antani, S., Thoma, G., McDonald, C.J.: Automatic screening for tuberculosis in chest radiographs: a survey. Quant. Imaging Med. Surg. 3(2), 89 (2013)
Woodring, J.: Recognition of pleural effusion in supine radiographs: how much fluid is required? Am. Roentgen Ray Soc. (AJR) 142, 59–64 (1984)
Avni, U., Greenspan, H., Konen, E., Sharon, M., Goldberger, J.: X-ray categorization and retrieval on the organ and pathology level, using patch-based visual words. IEEE Trans. Med. Imaging 30(3), 733–746 (2011)
Maduskar, P., Philipsen, R.H.M.M., Melendez, J., Scholten, E., Chanda, D., Ayles, H., Snchez, C.I., van Ginneken, B.: Automatic detection of pleural effusion in chest radiographs. Med. Image Anal. 28, 22–32 (2016)
Candemir, S., Jaeger, S., Palaniappan, K., Musco, J.P., Singh, R.K., Xue, Z., Karargyris, A., Antani, S., Thoma, G., McDonald, C.J.: Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration. IEEE Trans. Med. Imaging 33(2), 577–590 (2014)
Breiman, L.: Random forests. Mach. Learn. 45(1), 5–32 (2001)
MATLAB (R2016a), MathWorks, Natick, MA (2016)
Jaeger, S., Candemir, S., Antani, S., Wang, Y.-X.J., Pu-Xuan, L., Thoma, G.: Two public chest X-ray datasets for computer-aided screening of pulmonary diseases. Quant. Imaging Med. Surg. 4(6), 475–477 (2014)
Acknowledgement
The authors would like to express their thanks to Dr. Anurag Agrawal, CSIR Institute of Genomics and Integrative Biology, and Dr. Anjali Agrawal, Teleradiology Solutions, for their help in describing the methods they use while reading CXRs and their help in annotating the data set with instances of PE.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Sharma, U., Lall, B. (2017). Computer Aided Diagnosis of Pleural Effusion in Tuberculosis Chest Radiographs. In: Battiato, S., Gallo, G., Schettini, R., Stanco, F. (eds) Image Analysis and Processing - ICIAP 2017 . ICIAP 2017. Lecture Notes in Computer Science(), vol 10484. Springer, Cham. https://doi.org/10.1007/978-3-319-68560-1_55
Download citation
DOI: https://doi.org/10.1007/978-3-319-68560-1_55
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-68559-5
Online ISBN: 978-3-319-68560-1
eBook Packages: Computer ScienceComputer Science (R0)