Abstract
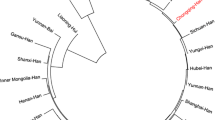
Forensic statistical parameters based on allelic frequencies of commonly used short tandem repeats were estimated for the Han population of Jiangsu province from P.R. China. The 6-dye GlobalFiler™ PCR amplification kit incorporates 21 autosomal STRs, providing reliable DNA typing results with enhanced the power of discrimination. Here, we analyzed the GlobalFiler™ STR loci in 516 unrelated individuals from Jiangsu Han population. A total of 256 alleles were observed ranging between 5 and 35.2 repeat units, and SE33 showed the greatest power of discrimination (34 alleles) in Jiangsu Han population. Most of the loci were found to be in the Hardy-Weinberg equilibrium after the Bonferroni correction with the exception of D3S1358. The combined power of exclusion (CPE) was 0.999999996353609, and the combined match probability (CMP) was 3.64 × 10−25. Phylogenetic parameters including pairwise genetic distances showed that Han population living in Jiangsu had closest genetic relationship with other East Asian populations. The present study provides precise reference database for forensic applications and population genetic studies.

Similar content being viewed by others
References
Mark ML, Savige G, Wattanapenpaiboon N (2004) Cuisine and health: a new initiative for science and technology “the Zhejiang report” from Hangzhou. Asia Pac J Clin Nutr 13:121–124
Hammond HA, Jin L, Zhong Y, Caskey CT, Chakraborty R (1994) Evaluation of 13 short tandem repeat loci for use in personal identification applications. Am J Hum Genet 55:175–189
Gettings KB, Aponte RA, Vallone PM, Butler JM (2015) STR allele sequence variation: current knowledge and future issues. Forensic Sci Int Genet 18:118–130. https://doi.org/10.1016/j.fsigen.2015.06.005
Wang DY, Gopinath S, Lagacé RE, Norona W, Hennessy LK, Short ML, Mulero JJ (2015) Developmental validation of the GlobalFiler(®) express PCR amplification kit: a 6-dye multiplex assay for the direct amplification of reference samples. Forensic Sci Int Genet 19:148–155. https://doi.org/10.1016/j.fsigen.2015.07.013
Adnan A, Ralf A, Rakha A, Kousouri N, Kayser M (2016) Improving empirical evidence on differentiating closely related men with RM Y-STRs: a comprehensive pedigree study from Pakistan. Forensic Sci Int Genet 25:45–51. https://doi.org/10.1016/j.fsigen.2016.07.005
Bi R, Zhang W, Yu D, Li X, Wang HZ, Hu QX, Zhang C, Lu W, Ni J, Fang Y, Li T, Yao YG (2015) Mitochondrial DNA haplogroup B5 confers genetic susceptibility to Alzheimer’s disease in Han Chinese. Neurobiol Aging 36:1604.e7–1604.16. https://doi.org/10.1016/j.neurobiolaging.2014.10.009
Yao J, Xing J, Xuan J, Wang B (2016) Population data of 15 autosomal STR loci in Chinese Han population from Jiangsu Province, Eastern China. Forensic Sci Int Genet 24:112–113. https://doi.org/10.1016/j.fsigen.2016.06.015
Yao J, Xiong K-Y, Zhang Y (2017) Population data of 19 autosomal STR loci in the Chinese Han population from Jiujiang, Southern China. Forensic Sci Int Genet 28:e47–e48. https://doi.org/10.1016/j.fsigen.2017.03.007
Bär W, Brinkmann B, Budowle B, Carracedo A, Gill P, Lincoln P, Mayr W, Olaisen B (1997) DNA recommendations. Further report of the DNA Commission of the ISFH regarding the use of short tandem repeat systems. International Society for Forensic Haemogenetics. Int J Legal Med 110:175–176
Gusmão L, Butler JM, Carracedo A, Gill P, Kayser M, Mayr WR, Morling N, Prinz M, Roewer L, Tyler-Smith C, Schneider PM, DNA Commission of the International Society of Forensic Genetics (2006) DNA Commission of the International Society of Forensic Genetics (ISFG): an update of the recommendations on the use of Y-STRs in forensic analysis. Forensic Sci Int 157:187–197. https://doi.org/10.1016/j.forsciint.2005.04.002
Bodner M, Bastisch I, Butler JM, Fimmers R, Gill P, Gusmão L, Morling N, Phillips C, Prinz M, Schneider PM, Parson W (2016) Recommendations of the DNA Commission of the International Society for Forensic Genetics (ISFG) on quality control of autosomal Short Tandem Repeat allele frequency databasing (STRidER). Forensic Sci Int Genet 24:97–102. https://doi.org/10.1016/j.fsigen.2016.06.008
Wu L, Pei B, Ran P, Song X (2017) Population genetic analysis of Xiamen Han population on 21 short tandem repeat loci. Legal Med 26:41–44. https://doi.org/10.1016/j.legalmed.2017.03.002
Li X, Li L, Wang Q, Zhang J, Ge W, Bai R, Yu X, Shi M (2017) Population genetic analysis of the Globalfiler STR loci in 3032 individuals from the Altay Han population of Xinjiang in northwest China. Int J Legal Med 132:141–143. https://doi.org/10.1007/s00414-017-1641-3
Park H-C, Kim K, Nam Y, Park J, Lee J, Lee H, Kwon H, Jin H, Kim W, Kim W, Lim S (2016) Population genetic study for 24 STR loci and Y indel (GlobalFiler™ PCR Amplification kit and PowerPlex® Fusion system) in 1000 Korean individuals. Legal Med 21:53–57. https://doi.org/10.1016/j.legalmed.2016.06.003
Fujii K, Watahiki H, Mita Y, Iwashima Y, Kitayama T, Nakahara H, Mizuno N, Sekiguchi K (2015) Allele frequencies for 21 autosomal short tandem repeat loci obtained using GlobalFiler in a sample of 1501 individuals from the Japanese population. Legal Med 17:306–308. https://doi.org/10.1016/j.legalmed.2015.08.007
Choi E-J, Park K-W, Lee Y-H, Nam YH, Suren G, Ganbold U, Kim JA, Kim SY, Kim HM, Kim K, Kim W (2017) Forensic and population genetic analyses of the GlobalFiler STR loci in the Mongolian population. Genes Genomics 39:423–431. https://doi.org/10.1007/s13258-016-0511-6
Alsafiah HM, Goodwin WH, Hadi S, Alshaikhi MA, Wepeba PP (2017) Population genetic data for 21 autosomal STR loci for the Saudi Arabian population using the GlobalFiler ® PCR amplification kit. Forensic Sci Int Genet 31:e59–e61. https://doi.org/10.1016/j.fsigen.2017.09.014
Zhang H, Yang S, Guo W, Ren B, Pu L, Ma T, Xia M, Jin L, Li L, Li S (2016) Population genetic analysis of the GlobalFiler STR loci in 748 individuals from the Kazakh population of Xinjiang in northwest China. Int J Legal Med 130:1187–1189. https://doi.org/10.1007/s00414-016-1319-2
Zhang H, Xia M, Qi L, Dong L, Song S, Ma T, Yang S, Jin L, Li L, Li S (2016) Forensic and population genetic analysis of Xinjiang Uyghur population on 21 short tandem repeat loci of 6-dye GlobalFiler™ PCR Amplification kit. Forensic Sci Int Genet 22:22–24. https://doi.org/10.1016/j.fsigen.2016.01.005
Felsenstein J (2009) PHYLIP (phylogeny inference package) Version 3.69. Department of Genome Sciences, University of Washington, Seattle
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Funding
This project is supported by the National Natural Science Foundation of P.R. China (NSFC, No. 81471826), Ministry of Finance, P.R. China.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
All participants gave their informed consent in writing after the study aims and procedures were carefully explained to them in their own language. The study was approved by the ethical review board of the China Medical University, Shenyang Liaoning Province, People’s Republic of China, and in accordance with the standards of the Declaration of Helsinki.
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Supplementary Table 1
Allele frequencies and forensic parameters derived from 21 autosomal STR loci for the Han population from Jiangsu Province. (XLSX 138 kb)
Supplementary Table 2
p-values from exact tests for linkage equilibrium (LE) employing pairwise combinations of all 21 STR loci in Han population from Jiangsu Province. (XLSX 10 kb)
Supplementary Table 3
Linkage disequilibrium test between loci located on the same chromosome in Han population from Jiangsu Province (XLSX 8 kb)
Supplementary Table 4
Genetic distance (FST) and associated p-values for population differentiation between the Uzbek population and other published populations (XLSX 13 kb)
Supplementary Table 5
Nei’s pairwise genetic distance between Han population from Jiangsu Province of China and other populations at the 21 Globalfiler autosomal STR loci (XLSX 8 kb)
Rights and permissions
About this article
Cite this article
Adnan, A., Zhan, X., Kasim, K. et al. Population data and phylogenetic structure of Han population from Jiangsu province of China on GlobalFiler STR loci. Int J Legal Med 132, 1301–1304 (2018). https://doi.org/10.1007/s00414-018-1815-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-018-1815-7