Abstract
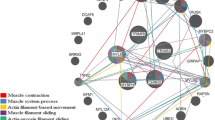
Beef tenderness contributes significantly to variation of beef palatability, and is largely influenced by various genetic and environmental factors. To identify candidate genes and pathways related to beef tenderness, we analyzed the longissimus dorsi (LD) muscle of Angus cattle that had different degrees of tenderness, measured by Warner–Bratzler shear force (WBSF). Microarray and RT-PCR analyses identified 53 genes that were differentially expressed in LD samples categorized as either tough or tender, including myosin, heavy chain 3 skeletal muscle embryonic (MYH3), myosin heavy chain 8 skeletal muscle perinatal (MYH8), guanylate binding protein 5 (GBP5), fatty acid binding protein 4 (FABP4), Stearoyl-coenzyme A desaturase (SCD), Fatty acid synthase (FASN), ubiquitin-like with PHD and ring finger domains 1 (UHRF1). Most of these genes are involved in lipid metabolism and skeletal muscle contraction. Employing Gene ontology (GO) and Ingenuity Pathway Analysis (IPA), several GO terms and pathways were found to be related to hydrolase, peptidase and GTPase activity, lipid metabolism, small molecule biochemistry, molecular transport, and tissue development. Overall, this analysis provides insight into the metabolic relationships between muscle biology and beef quality.





Similar content being viewed by others
References
Boleman SJ, Boleman SL, Miller RK, Taylor JF, Cross HR, Wheeler TL, Koohmaraie M, Shackelford SD, Miller MF, West RL, Johnson DD, Savell JW (1997) Consumer evaluation of beef of known categories of tenderness. J Anim Sci 75:1521–1524
Goodson KJ, Morgan WW, Reagan JO, Gwartney BL, Courington SM, Wise JW, Savell JW (2002) Beef customer satisfaction: factors affecting consumer evaluations of clod steaks. J Anim Sci 80:401–408
Brady DE (1937) A study of the factors influencing tenderness and texture of beef. J Anim Sci 1937:246–250
Hertzman C, Olsson U, Tornberg E (1993) The influence of high temperature, type of muscle and electrical stimulation on the course of rigor, ageing and tenderness of beef muscles. Meat Sci 35:119–141
Minks D, Stringer WC (1972) The influence of aging beef in vacuum. J Food Sci 37:736–738
Monsón F, Sañudo C, Sierra I (2005) Influence of breed and ageing time on the sensory meat quality and consumer acceptability in intensively reared beef. Meat Sci 71:471–479
Neely TR, Lorenzen CL, Miller RK, Tatum JD, Wise JW, Taylor JF, Buyck MJ, Reagan JO, Savell JW (1999) Beef customer satisfaction: cooking method and degree of doneness effects on the top round steak. J Anim Sci 77:653–660
O’Connor SF, Tatum JD, Wulf DM, Green RD, Smith GC (1997) Genetic effects on beef tenderness in Bos indicus composite and Bos taurus cattle. J Anim Sci 75:1822–1830
Koohmaraie M, Geesink GH (2006) Contribution of postmortem muscle biochemistry to the delivery of consistent meat quality with particular focus on the calpain system. Meat Sci 74:34–43
Nishimura T, Hattori A, Takahashi K (1999) Structural changes in intramuscular connective tissue during the fattening of Japanese Black cattle: effect of marbling on beef tenderization. J Anim Sci 77:93–104
Tornberg E (1996) Biophysical aspects of meat tenderness. Meat Sci 43:175–191
Thompson JM, Perry D, Daly B, Gardner GE, Johnston DJ, Pethick DW (2006) Genetic and environmental effects on the muscle structure response post-mortem. Meat Sci 74:59–65
Huffman KL, Miller MF, Hoover LC, Wu CK, Brittin HC, Ramsey CB (1996) Effect of beef tenderness on consumer satisfaction with steaks consumed in the home and restaurant. J Anim Sci 74:91–97
Barendse W, Harrison BE, Bunch RJ, Thomas MB (2008) Variation at the Calpain 3 gene is associated with meat tenderness in zebu and composite breeds of cattle. BMC Genet 9:41
Davis GP, Moore SS, Drinkwater RD, Shorthose WR, Loxton ID, Barendse W, Hetzel DJS (2008) QTL for meat tenderness in the M. longissimus lumborum of cattle. Anim Genet 39:40–45
Gao Y, Zhang R, Hu X, Li N (2007) Application of genomic technologies to the improvement of meat quality of farm animals. Meat Sci 77:36–45
Hocquette JF, Renard G, Levéziel H, Picard B, Cassar-Malek I (2006) The potential benefits of genetics and genomics to improve beef quality—a review. Anim Sci Pap Rep 24:173–186
Jennifer G, Stephen B, Caroline MC, John W, Pamela W (2009) Association of selected SNP with carcass and taste panel assessed meat quality traits in a commercial population of Aberdeen Angus-sired beef cattle. Genet Sel Evol 41:36
Morzel M, Terlouw C, Chambon C, Micol D, Picard B (2008) Muscle proteome and meat eating qualities of longissimus thoracis of Blonde d’Aquitaine young bulls: a central role of HSP27 isoforms. Meat Sci 78:297–304
Mullen AM, Stapleton PC, Corcoran D, Hamill RM, White A (2006) Understanding meat quality through the application of genomic and proteomic approaches. Meat Sci 74:3–16
Koohmaraie M, Kent MP, Shackelford SD, Veiseth E, Wheeler TL (2002) Meat tenderness and muscle growth: is there any relationship? Meat Sci 62:345–352
Chen FY, Niu H, Wang JQ, Lei CZ, Lan XY, Zhang CL, Li MJ, Hua LS, Wang J, Chen H (2011) Polymorphism of DLK1 and CLPG gene and their association with phenotypic traits in Chinese cattle. Mol Biol Rep 38:243–248
Bernard C, Cassar-Malek I, Le Cunff M, Dubroeucq H, Renand G, Hocquette JF (2007) New indicators of beef sensory quality revealed by expression of specific genes. J Agric Food Chem 55:5229–5237
Iglesias PP, Caffaro ME, Amadio AF, Arias Mañotti A, Poli MA (2011) CAPN1 markers in three Argentinean cattle breeds: report of a new InDel polymorphism within intron 17. Mol Biol Rep 38:1645–1649
Iwanowska A, Grześ B, Mikołajczak B, Iwańska E, Juszczuk-Kubiak E, Rosochacki SJ, Pospiech E (2011) Impact of polymorphism of the regulatory subunit of the -calpain (CAPN1S) on the proteolysis process and meat tenderness of young cattle. Mol Biol Rep 38:1295–1300
Fan YY, Zan LS, Fu CZ, Tian WQ, Wang HB, Liu YY, Xin YP (2011) Three novel SNPs in the coding region of PPAR gene and their associations with meat quality traits in cattle. Mol Biol Rep 38:131–137
Zhang YY, Zan LS, Wang HB (2011) Screening candidate genes related to tenderness trait in Qinchuan cattle by genome array. Mol Biol Rep 38:2007–2014
Zapata I, Zerby HN, Wick M (2009) Functional proteomic analysis predicts beef tenderness and the tenderness differential. J Agric Food Chem 57:4956–4963
Sawdy JC, Kaiser SA, St-Pierre NR, Wick MP (2004) Myofibrillar 1-D fingerprints and myosin heavy chain MS analyses of beef loin at 36 h postmortem correlate with tenderness at 7 days. Meat Sci 67:421–426
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95:14863
Zheng Q, Wang XJ (2008) GOEAST: a web-based software toolkit for Gene Ontology Enrichment Analysis. Nucleic Acids Res 36:358–363
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408
Wang YH, Reverter A, Tan SH, De Jager N, Wang R, McWilliam SM, Cafe LM, Greenwood PL, Lehnert SA (2009) Gene expression patterns during intramuscular fat development in cattle. J Anim Sci 87:119–130
Perry D, Shorthose WR, Ferguson DM, Thompson JM (2001) Methods used in the CRC program for the determination of carcass yield and beef quality. Aust J Exp Agric 41:953–1040
Hellerstein MK, Christiansen M, Kaempfer S, Kletke C, Wu K, Reid JS, Mulligan K, Hellerstein NS, Shackleton CH (1991) Measurement of de novo hepatic lipogenesis in humans using stable isotopes. J Clin Invest 87:1841
El-Kadi SW, Baldwin RL, Sunny NE, Owens SL, Bequette BJ (2006) Intestinal protein supply alters amino acid, but not glucose, metabolism by the sheep gastrointestinal tract. J Nutr 136:1261
Boleman SJ, Boleman SL, Miller RK, Taylor JF, Cross HR, Wheeler TL, Koohmaraie M, Shackelford SD, Miller MF, West RL (1997) Consumer evaluation of beef of known categories of tenderness. J Anim Sci 75:1521–1524
Behrends JM, Goodson KJ, Koohmaraie M, Shackelford SD, Wheeler TL, Morgan WW, Reagan JO, Gwartney BL, Wise JW, Savell JW (2005) Beef customer satisfaction: USDA quality grade and marination effects on consumer evaluations of top round steaks. J Anim Sci 83:662–670
Wheeler TL, Shackelford SD, Koohmaraie M (1999) Tenderness classification of beef: IV. Effect of USDA quality grade on the palatability of “tender” beef longissimus when cooked well done. J Anim Sci 77:882–888
George MH, Tatum JD, Dolezal HG, Morgan JB, Wise JW, Calkins CR, Gordon T, Reagan JO, Smith GC (1997) Comparison of USDA quality grade with Tendertec for the assessment of beef palatability. J Anim Sci 75:1538–1546
Miller MF, Kerth CR, Wise JW, Lansdell JL, Stowell JE, Ramsey CB (1997) Slaughter plant location, USDA quality grade, external fat thickness, and aging time effects on sensory characteristics of beef loin strip steak. J Anim Sci 75:662–667
Jeremiah LE (1970) Beef quality. I. Marblingas an indicator of palatability. Texas Agricultural Experiment Station Technical Bulletin 22, College Station, TX
DeVol DL, Mckeith FK, Bechtel PJ, Novakofski J, Shanks RD, Carr TR (1988) Variation on composition and palatability traits and relationships between muscle characteristics abd palatability in a random sample of pork carcasses. J Anim Sci 66:385–395
Jeremiah LE (1983) The influence of inherent muscle quality upon the cooking losses from and palatability attributes of pork loin chops. In: Proceedings of Annual Meeting of the Western Section of the American Society of Animal Science, vol 34, pp 109–111
Hoashi S, Hinenoya T, Tanaka A, Ohsaki H, Sasazaki S, Taniguchi M, Oyama K, Mukai F, Mannen H (2008) Association between fatty acid compositions and genotypes of FABP 4 and LXR-alpha in Japanese Black cattle. BMC Genet 9:84
Yang A, Larsen TW, Smith SB, Tume RK (1999) 9 desaturase activity in bovine subcutaneous adipose tissue of different fatty acid composition desaturase activity in bovine subcutaneous adipose tissue of different fatty acid composition. Lipids 34:971–978
Zhang L, Ge L, Parimoo S, Stenn K, Prouty SM (1999) Human stearoyl-CoA desaturase: alternative transcripts generated from a single gene by usage of tandem polyadenylation sites. Biochem J 340:255–264
Li J, Ding SF, Habib NA, Fermor BF, Wood CB, Gilmour RS (1994) Partial characterization of a cDNA for human stearoyl CoA desaturase and changes in its mRNA expression in some normal and malignant tissues. Int J Cancer 57:348–352
Jayakumar A, Tai MH, Huang WY, Al-Feel W, Hsu M, Abu-Elheiga L, Chirala SS, Wakil SJ (1995) Human fatty acid synthase: properties and molecular cloning. Proc Natl Acad Sci USA 92:8695–8699
Chakravarty B, Gu Z, Chirala SS, Wakil SJ, Quiocho FA (2004) Human fatty acid synthase: structure and substrate selectivity of the thioesterase domain. Proc Natl Acad Sci USA 101:15567–15572
Mannen H (2011) Identification and utilization of genes associated with beef qualities. Anim Sci J 82:1–7
Roy R, Ordovas L, Zaragoza P, Romero A, Moreno C, Altarriba J, Rodellar C (2006) Association of polymorphisms in the bovine FASN gene with milk-fat content. Anim Genet 37:215–218
Huff-Lonergan E, Baas TJ, Malek M, Dekkers JC, Prusa K, Rothschild MF (2002) Correlations among selected pork quality traits. J Anim Sci 80:617–627
Bouley J, Chambon C, Picard B (2004) Mapping of bovine skeletal muscle proteins using two dimensional gel electrophoresis and mass spectrometry. Proteomics 4:1811–1824
Lubeseder-Martellato C, Guenzi E, Jörg A, Töpolt K, Naschberger E, Kremmer E, Zietz C, Tschachler E, Hutzler P, Schwemmle M (2002) Guanylate-binding protein-1 expression is selectively induced by inflammatory cytokines and is an activation marker of endothelial cells during inflammatory diseases. Am J Pathol 161:1749–1759
Guenzi E, Töpolt K, Lubeseder-Martellato C, Jörg A, Naschberger E, Benelli R, Albini A, Stürzl M (2003) The guanylate binding protein-1 GTPase controls the invasive and angiogenic capability of endothelial cells through inhibition of MMP-1 expression. EMBO J 22:3772–3782
Lu K, Lee MH, Hazard S, Brooks-Wilson A, Hidaka H, Kojima H, Ose L, Stalenhoef AFH, Mietinnen T, Bjorkhem I (2001) Two genes that map to the STSL locus cause sitosterolemia: genomic structure and spectrum of mutations involving sterolin-1 and sterolin-2, encoded by ABCG5 and ABCG8, respectively. Am J Hum Genet 69:278–290
Farke C, Meyer HHD, Bruckmaier RM, Albrecht C (2008) Differential expression of ABC transporters and their regulatory genes during lactation and dry period in bovine mammary tissue. J Dairy Res 75:406–414
Viturro E, Farke C, Meyer HHD, Albrecht C (2006) Identification, sequence analysis and mRNA tissue distribution of the bovine sterol transporters ABCG5 and ABCG8. J Dairy Sci 89:553–561
Acknowledgments
The work was supported by China Scholarship Council (CSC), Maryland Agricultural Experiment Station (MAES) and Jorgensen Endowment Funds.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, C., Tian, F., Yu, Y. et al. Muscle transcriptomic analyses in Angus cattle with divergent tenderness. Mol Biol Rep 39, 4185–4193 (2012). https://doi.org/10.1007/s11033-011-1203-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-1203-6