Abstract
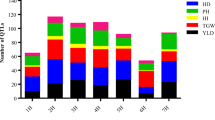
Several reports on mapping and introgression of quantitative trait loci (QTLs) for yield and related traits from wild species showed their importance in yield improvement. The aim of this study was to locate common major effect, consistent and precise yield QTLs across the wild species of rice by applying genome-wide QTL meta-analysis for their use in marker-aided selection (MAS) and candidate gene identification. Seventy-six yield QTLs reported in 11 studies involving inter-specific crosses were projected on a consensus map consisting of 699 markers. The integration of 11 maps resulted in a consensuses map of 1,676 cM. The number of markers ranged from 32 on chromosome 12 to 96 on chromosome 1. The order of markers between consensus map and original map was generally consistent. Meta-analysis of 68 yield QTLs resulted in 23 independent meta-QTLs on ten different chromosomes. Eight meta-QTLs were less than 1.3 Mb. The smallest confidence interval of a meta-QTL (MQTL) was 179.6 kb. Four MQTLs were around 500 kb and two of these correspond to a reasonably small genetic distance 4.6 and 5.2 cM, respectively, and suitable for MAS. MQTL8.2 was 326-kb long with a 35-cM interval indicating it was in a recombination hot spot and suitable for fine mapping. Our results demonstrate the narrowing down of initial yield QTLs by Meta-analysis and thus enabling short listing of QTLs worthy of MAS or fine mapping. The candidate genes shortlisted are useful in validating their function either by loss of function or over expression.







Similar content being viewed by others
References
Akagi H, Nakamura A, MisonoY IA, Takahashi H, Mori K, Fujimura T (2004) Positional cloning of the rice Rf-1 gene, a restorer of BT-type cytoplasmic male sterility that encodes a mitochondria targeting PPR protein. Theor Appl Genet 108:1449–1457
Akaike H (1974) A new look at the statistical model identification. IEEE Trans Autom Control 19:716–723
Arcade A, Labourdette A, Falque M, Mangin B, Chardon F, Charcosset A, Joets J (2004) BioMercator: integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics 14:2324–2326
Ashikari M, Matsuoka M (2006) Identification, isolation and pyramiding of quantitative trait loci for rice breeding. Trends Plant Sci 11:344–350
Ashikari M, Sakakibara H, Lin S, Ymamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M (2005) Cytokinin oxidase regulates rice grain production. Science 309:741–745
Ballini E, Morel JB, Droc G, Price AH, Courtois B, Nottehem JL, Tharreau DA (2008) Genome wide meta analysis of rice blast resistance genes and quantitative trait loci provides new insights into partial and complete resistance. Mol Plant Microbe Intera 21:859–868
Bentolila S, Alfonso AA, Hanson MR (2002) A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc Natl Acad Sci 99:10887–10892
Brondani C, Rangel PHN, Brondani RPV, Ferreira ME (2002) QTL mapping and introgression of yield-related traits from Oryza glumaepatula to cultivated rice (Oryza sativa) using microsatellite markers. Theor Appl Genet 104:1192–1203
Chardon F, Virlon B, Moreau L, Falque M, Joets J, Decousset L, Murigneux A, Charcosset A (2004) Genetic architecture of flowering time in maize as inferred from quantitative trait loci meta-analysis and synteny conservation with rice genome. Genetics 168:2169–2185
Courtois B, Ahmadi N, Khowaja F, Price AH, Rami J, Frouin J, Hamelin C, Ruiz M (2009) Rice root genetic architecture: meta-analysis from a drought QTL database. Rice 2:115–128
Darvasi A, Soller M (1997) A simple method to calculate resolving power and confidence interval of QTL map location. Behav Genet 27:125–132
Dong ZY, Wang YM, Zhang ZJ, Shen Y, Lin XY, Ou XF, Han FP, Liu B (2006) Extent and pattern of DNA methylation alteration in rice lines derived from introgressive hybridization of rice and Zizania latifolia Griseb. Theor Appl Genet 113:196–205
Fridman E, Carrari F, Liu YS, Fernie AR, Zamir D (2004) Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science 305:1786–1789
Fu J, Keurentjes JJ, Bouwmeester H, America T, Verstappen FW, Ward JL, Beale MH, Devos RC, Dijkstra M, Scheltema RA, Johannes F, Koornneef M, Vreugdenhil D, Breitling R, Jansen RC (2009) System-wide molecular evidence for phenotypic buffering in Arabidopsis. Nat Genet 4:144–145
Goffinet B, Gerber S (2000) Quantitative trait loci: a meta-analysis. Genetics 155:463–473
Guo B, Sleper DA, Lu P, Shannon JG, Nguyen HT, Arelli PR (2006) QTLs associated with resistance to soybean cyst nematode in soybean meta-analysis of QTL locations. Crop Sci 46:202–208
Gur A, Zamir D (2004) Unused natural variation can lift yield barriers in plant breeding. PLoS Biol 2:e245
Hanocq E, Laperche A, Jaminon O, Laine AL, Gouis JL (2007) Most significant genome regions involved in the control of earliness traits in bread wheat, as revealed by QTL meta-analysis. Theor Appl Genet 114:569–584
Hao Z, Li X, Liu X, Xie C, Li M, Zhang D, Zhang S (2010) Meta-analysis of constitutive and adaptive QTL for drought tolerance in maize. Euphytica 174:165–177
Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP (2007) F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol 143:1467–1483
Kaladhar K (2006) Mapping of Quantitative Trait Loci (QTL) for yield and related traits in BC2F2 population of O. sativa cv Swarna × O. nivara (IRGC81832). PhD thesis Osmania University, Hyderabad, India
Kovach MJ, McCouch SR (2008) Leveraging natural diversity: back through the bottleneck. Curr Opin Plant Biotechnol 11:193–200
Lanaud C, Fouet O, Clement D, Boccara M, Risterucci AM, Maharaj SS, Legavre T, Argout X (2009) A meta-QTL analysis of disease resistance traits of Theobroma cacao L. Mol Breed 24:361–374
Li C, Zhou A, Sang T (2006) Genetic analysis of rice domestication syndrome with the wild annual species, Oryza nivara. New Phytol 170:185–194
Liang F, Deng Q, Wang Y, Xiong Y, Jin D, Li J, Wang B (2004) Molecular marker assisted selection for yield enhancing genes in the progeny of “9311 × O. rufipogon” using SSR. Euphytica 139:159–165
Lin Y, Schertz K, Paterson A (1995) Comparative analysis of QTLs affecting plant height and maturity across the Poaceae, in reference to an interspecific sorghum population. Genetics 141:391–411
Loffler M, Schon CC, Miedaner T (2009) Revealing the genetic architecture of FHB resistance in hexaploid wheat (Triticum aestivum L.) by QTL meta-analysis. Mol Breed 23:473–488
Marri PR, Sarla N, Reddy LV, Siddiq EA (2005) Identification and mapping of yield and related QTLs from an Indian accession of O. rufipogon. BMC Genet 6:33
McCouch SR, Sweeney M, Li J, Jiang H, Thomson M, Septiningsih E, Edwards J, Moncada P, Xiao J, Garris A, Tai T, Martinez C, Tohme J, Sugiono M, McClung A, Yuan LP, Ahn SN (2007) Through the genetic bottleneck: O. rufipogon as a source of trait-enhancing alleles for O. sativa. Euphytica 154:317–339
McCouch et al (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res 9:199–207
Moncada P, Martinez CP, Borrero J, Chatel M, Gouch JH, Glumaraes E, Tohme J, Mc Couch SR (2001) Qualitative trait loci for yield and yield components in an Oryza sativa × Oryza rufipogon BC2F2 population evaluated in an upland environment. Theor Appl Genet 102:41–52
Mukhopadhyay A, Vij S, Tyagi AK (2004) Over expression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc Natl Acad Sci 101:6309–6314
Price AH (2006) Believe it or not, QTLs are accurate. Trends Plant Sci 11:213–216
Rahman ML, Chu SH, Choi M, Qiao YL, Jiang W, Piao R, Khanam S, Cho Y, Jeung J, Jena KK, Koh H (2008) Identification of QTLs for some agronomic traits in rice using an introgression line from Oryza minuta. Mol Cells 24:16–26
Rong J, Feltus FA, Waghmare VN, Pierce GJ, Chee PW, Draye X, Saranga Y, Wright RJ, Wilkins TA, May OL, Smith CW, Gannaway JR, Wendel JF, Paterson AH (2007) Meta-analysis of polyploid cotton QTL shows unequal contributions of subgenomes to a complex network of genes and gene clusters implicated in lint fiber development. Genetics 176:2577–2588
Sakamoto T, Morinaka Y, Ohnishi T, Sunohara H, Fujioka S, Tanaka MU, Mizutani M, Sakata K, Takatsuto S, Yoshida S, Tanaka H, Kitano H, Matsuoka M (2005) Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotechnol 24:105–109
Salvi S, Tuberosa R (2005) To clone or not to clone plant QTLs: present and future challenges. Trends Plant Sci 10:297–373
Septiningsih EM, Prasetiyono J, Lubis E, Tai TH, Tjubaryat T, Moeljopawiro S, McCouch SR (2003) Identification of quantitative trait loci for yield and yield components in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O. rufipogon. Theor Appl Genet 107:1419–1432
Swamy BPM (2008) Genome wide mapping of Quantitaive Trait Loci for yield and grain quality traits in O. sativa cv Swarna × O. nivara (IRGC81848) population. Phd thesis Osmania University, Hyderabad, India
Swamy BPM, Sarla N (2008) Yield enhancing QTLs from wild species. Biotechnol Adv 26:106–120
Tan L, Liu F, Xue W, Wang G, Ye S, Zhu Z, Fu Y, Wang X, Sun C (2007) Development of Oryza rufipogon and O. sativa introgression lines and assessment for yield-related quantitative trait loci. J Integr Plant Biol 49:871–884
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch SR (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations and genetic marker potential. Genome Res 11:1441–1452
Thomson MJ, Tai TH, McClung AM, Lai XH, Hinga ME, Lobos KB, Xu Y, Martinez CP, McCouch SR (2003) Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor Appl Genet 107:479–493
Tian F, Li DJ, Fu Q, Zhu ZF, Fu YC, Wang XK, Sun CQ (2006) Construction of introgression lines carrying wild rice (Oryza rufipogon Griff) segments in cultivated rice (Oryza sativa L.) background and characterization of introgressed segments associated with yield-related traits. Theor Appl Genet 112:570–580
Wang YM, Dong ZY, Zhang ZJ, Lin XY, Shen Y, Zhou D, Liu B (2005) Extensive denovo variation in rice induced by introgression from wild rice (Zizania latifolia). Genetics 170:1945–1956
Wang L, Xu Y, Zhang C, Ma Q, Joo SH, Kim SK, Xu Z, Chong K (2008) OsLIC, a Novel CCCH-type zinc finger protein with transcription activation, mediates rice architecture via brassinosteroids signaling. PLoS ONE 3:1–12
Wang K, Feng G, Zhu R, Li S, Zhu Y (2010) Expression, purification, and secondary structure prediction of pentatricopeptide repeat protein RF1A from rice. Plant Mol Biol Rep. doi:10.1007/s11105-010-0260-7
Xiao J, Li J, Grandillo S, Ahn SN, Yuan L, Steven D, McCouch SR (1998) Identification of trait-improving quantitative trait loci alleles from a wild rice relative, Oryza rufipogon. Genetics 150:899–909
Xin M, Feng D, Wang H, Li X, Kong L (2010) Cloning and expression analysis of wheat cytokinin oxidase/dehydrogenase gene TaCKX3. Plant Mol Biol Rep. doi:10.1007/s11105-010-0209
Xu K, Xu X, Fukao T, Canlas P, Rodriguez RM, Heuer S, Ismail AM, Serres JB, Ronald PC, Mackill DJ (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442:705–708
Xu X, Liu Z, Zhang D, Liu Y, Song W, Li J, Dai J (2009) Isolation and analysis of rice Rf1-orthologus PPR genes co-segregating with Rf3 in maize. Plant Mol Biol Rep 27:511–517
Yoon DB, Kang KH, Kim HJ, Ju HG, Kwon SJ, Suh JP, Jeong OY, Ahn SN (2006) Mapping quantitative trait loci for yield components and morphological traits in an advanced backcross population between Oryza grandiglumis and the O. sativa japonica cultivar Hwaseongbyeo. Theor Appl Genet 112:1052–1062
Yu S, Li J, Luo L (2010) Complexity and specificity of precursor microRNAs driven by transposable elements in rice. Plant Mol Biol Rep 28:502–511
Zhang X, Guo X, Lei C, Cheng Z, Lin Q, Wang J, Wu F, Wang J, Wan J (2010) Over expression of SlCZFP1, a novel TFIIIA-type zinc finger protein from tomato, confers enhanced cold tolerance in transgenic Arabidopsis and rice. Plant Mol Biol Rep. doi:10.1007/s11105-010-0223
Acknowledgment
BPMS thanks UGC–CSIR for Senior Research Fellowship. NS thanks Department of Biotechnology, Government of India for financial support to the Network Project on Functional Genomics of rice at DRR. We thank the Director of DRR for constant support and encouragement.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Table 1
(DOC 166 kb)
Rights and permissions
About this article
Cite this article
Swamy, B.P.M., Sarla, N. Meta-analysis of Yield QTLs Derived from Inter-specific Crosses of Rice Reveals Consensus Regions and Candidate Genes. Plant Mol Biol Rep 29, 663–680 (2011). https://doi.org/10.1007/s11105-010-0274-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-010-0274-1