Abstract
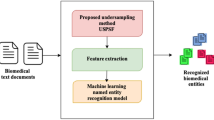
Named entity recognition (NER), as one of the crucial tasks of information extraction (IE), has important effect on the quality of its subsequent applications such as answering the question, co-reference resolution, relation discovery, etc. NER can be considered as a kind of classification problem, which has to deal with its own challenging issues. Class-Imbalanced Problem (CIP) is one of the important problems in classification domain from which almost all NER tasks also suffer, because usually, the number of entity mentions of interest in the given text is much less than undesired entities. The quality of the IE’s subtasks for which NER is the basis is directly affected by any improvement on the performances of NER systems. In this research, an effort has been made to increase the overall performance of NER systems by decreasing the curse of CIP as much as possible. A new heuristic approach based on the genetic algorithm has been devised to undersample the training data which is used for NER. Regarding the fact that given training patterns for NER are of individual sentence forms, in the developed approach, this issue is considered as well and it was applied to individual sentences from training data. The proposed method has been applied on two different corpuses: CoNLL corpus from newswire domain and JNLPBA from biomedical context to see its impact on different type of contexts. By increasing the performance in terms of F-score for both data sets, our proposed method outperforms the baseline systems using original data. Furthermore, in comparison with random undersampling, it results in better outcomes. In addition, the effect of considering sentences of training data individually in sampling process and taking all of them together has been investigated.




Similar content being viewed by others
References
Westergaard, D., Stærfeldt, H.-H., Tønsberg, C., Jensen, L.J., Brunak, S.: Text mining of 15 million full-text scientific articles. bioRxiv, 162099 (2017). https://doi.org/10.1101/162099
Holzinger, A., Schantl, J., Schroettner, M., Seifert, C., Verspoor, K.: Biomedical text mining: state-of-the-art, open problems and future challenges. In: Holzinger, A., Jurisica, I. (eds.) Interactive Knowledge Discovery and Data Mining in Biomedical Informatics, Lecture Notes in Computer Science, vol. 8401,pp. 271–300. Springer, Berlin (2014). https://doi.org/10.1007/978-3-662-43968-5_16
Munkhdalai, T., Li, M., Batsuren, K., Park, H.A., Choi, N.H., Ryu, K.H.: Incorporating domain knowledge in chemical and biomedical named entity recognition with word representations. J. Cheminform. 7(1), S9 (2015)
Marrero, M., Urbano, J., Sánchez-Cuadrado, S., Morato, J., Gómez-Berbís, J.M.: Named entity recognition: fallacies, challenges and opportunities. Comput. Stand. Interfaces 35(5), 482–489 (2013)
Yang, Q., Wu, X.: 10 challenging problems in data mining research. Int. J. Inf. Technol. Decis. Mak. 5(04), 597–604 (2006)
Akkasi, A., Varoğlu, E., Dimililer, N.: Balanced undersampling: a novel sentencebased undersampling method to improve recognition of named entities in chemical and biomedical text. Appl. Intell. 1–14 (2017). https://doi.org/10.1007/s10489-017-0920-5
Tang, B., Feng, Y., Wang, X., Wu, Y., Zhang, Y., Jiang, M., Xu, H.: A comparison of conditional random fields and structured support vector machines for chemical entity recognition in biomedical literature. J. Cheminform. 7(S1), S8 (2015)
Nanni, L., Fantozzi, C., Lazzarini, N.: Coupling different methods for overcoming the class imbalance problem. Neurocomputing 158, 48–61 (2015)
Lemnaru, E. C.: Strategies for dealing with real world classification problems. Doctoral dissertation, Technical University of Cluj-Napoca (2012)
Japkowicz, N.: The class imbalance problem: significance and strategies. In: Proceedings of the 2000 International Conference on Artificial Intelligence (IC-AI’2000): Special Track on Inductive Learning Las Vegas, Nevada (2000)
He, H., Ma, Y. (eds.): Imbalanced Learning: Foundations, Algorithms, and Applications. Wiley, New York (2013)
Zhu, B., Baesens, B., vanden Broucke, S.K.: An empirical comparison of techniques for the class imbalance problem in churn prediction. Inf. Sci. 408, 84–99 (2017)
Longadge, R., Dongre, S.: Class imbalance problem in data mining review (2013). arXiv preprint arXiv:1305.1707
Chawla, N. V.: Data mining for imbalanced datasets: an overview. In: Data Mining and Knowledge Discovery Handbook, pp. 875–886. Springer (2009). https://doi.org/10.1007/978-0-387-09823-4_45
Tomek, I.: Two Modifications of CNN. IEEE Trans. Syst. Man Commun. SMC 6, 769–772 (1976)
Kumar, R.R., Viswanath, P., Bindu, C.S.: Nearest neighbor classifiers: a review. Int. J. Comput. Intell. Res. 13(2), 303–311 (2017)
Faris, H.: Neighborhood cleaning rules and particle swarm optimization for predicting customer churn behavior in telecom industry. Int. J. Adv. Sci. Technol. 68, 11–22 (2014)
Kubat, M., Matwin, S.: Addressing the curse of imbalanced training sets: one-sided selection. In: Proceedings of the Fourteenth International Conference on Machine Learning, pp. 179–186. Morgan Kaufmann (1997)
Chawla, N.V., Bowyer, K.W., Hall, L.O., Kegelmeyer, W.P.: SMOTE: synthetic minority over-sampling technique. J. Artif. Intell. Res. 16, 321–357 (2002)
Han, H., Wang, W. Y., Mao, B. H.: Borderline-SMOTE: a new over-sampling method in imbalanced data sets learning. In: Advances in Intelligent Computing, pp. 878–887. Springer, Berlin (2005). https://doi.org/10.1007/11538059_91
Lim, P., Goh, C.K., Tan, K.C.: Evolutionary cluster-based synthetic oversampling ensemble (eco-ensemble) for imbalance learning. IEEE Trans. Cybern. 47, 2850–2861 (2016)
Braytee, A., Liu, W., Kennedy, P.: A cost-sensitive learning strategy for feature extraction from imbalanced data. In: International Conference on Neural Information Processing, pp. 78–86. Springer International Publishing (2016). https://doi.org/10.1007/978-3-319-46675-0_9
Chawla, N. V., Lazarevic, A., Hall, L. O., Bowyer, K. W.: SMOTEBoost: Improving prediction of the minority class in boosting. In: Knowledge Discovery in Databases: PKDD 2003, pp. 107–119. Springer, Berlin (2003). https://doi.org/10.1007/978-3-540-39804-2_12
Williams, G., Chen, H.: Stratified over-sampling bagging method for random forests on imbalanced data. In: Intelligence and Security Informatics: 11th Pacific Asia workshop. PAISI 2016, Auckland, New Zealand, April 19, 2016, Proceedings, vol. 9650, p. 63. Springer (2016). https://doi.org/10.1007/978-3-319-31863-9_5
Ahachad, A., Álvarez-Pérez, L., Figueiras-Vidal, A.R.: Boosting ensembles with controlled emphasis intensity. Pattern Recognit. Lett. 88, 1–5 (2017)
Tomanek, K., Hahn, U.: Reducing class imbalance during active learning for named entity annotation. In: Proceedings of the Fifth International Conference on Knowledge Capture, pp. 105–112. ACM (2009). https://doi.org/10.1145/1597735.1597754
Gliozzo, A.M., Giuliano, C., Rinaldi, R.: Instance filtering for entity recognition. ACM SIGKDD Explor. Newsl. 7(1), 11–18 (2005)
Mitchell, M.: An Introduction to Genetic Algorithms. MIT Press: Cambridge, MA (1998)
Dasgupta, D., Michalewicz, Z. (eds.): Evolutionary Algorithms in Engineering Applications. Springer Science & Business Media, New York (2013)
http://www.obitko.com/tutorials/genetic-algorithms/crossover-mutation.php. Accessed 12 Aug 2017
Sang, E.F., Veenstra, J.: Representing text chunks. In: Proceedings of the Ninth Conference on European Chapter of the Association for Computational Linguistics, pp. 173–179. Association for Computational Linguistics (1999)
Powers, D.M.W.: Evaluation: from precision, recall and f-measure to roc, informedness, markedness & correlation. J. Mach. Learn. Technol. 2(1), 37–63 (2011)
Tjong Kim Sang, E.F., De Meulder, F.: Introduction to the CoNLL-2003 shared task: Language-independent named entity recognition. In CoNLL-2003 (2003)
Kim, J.D., Ohta, T., Tsuruoka, Y., Tateisi, Y., Collier, N.: Introduction to the bio-entity recognition task at JNLPBA. In: Proceedings of the International Joint Workshop on Natural Language Processing in Biomedicine and its Applications, pp. 70–75. Association for Computational Linguistics (2004)
McCallum, Andrew Kachites. MALLET: A Machine Learning for Language Toolkit (2002). http://mallet.cs.umass.edu. Accessed 5 Oct 2017
Akkasi, A., Varoğlu, E., Dimililer, N.: ChemTok: a new rule based tokenizer for chemical named entity recognition. BioMed Res. Int. 2016 (2016)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Akkasi, A. Sentence-based undersampling for named entity recognition using genetic algorithm. Iran J Comput Sci 1, 165–174 (2018). https://doi.org/10.1007/s42044-018-0014-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42044-018-0014-5