Abstract
When applied to 3D image reconstruction, conventional landmark-based registration methods tend to generate unnatural vertical structures due to inconsistencies between the employed model and the real tissue. This paper demonstrates a fully non-rigid image registration method for 3D image reconstruction which considers the spatial continuity and smoothness of each constituent part of the microstructures in the tissue. Corresponding landmarks are detected along the images, defining a set of trajectories, which are smoothed out in order to define a diffeomorphic mapping. The resulting reconstructed 3D image preserves the original tissue architecture, allowing the observation of fine details and structures.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others

1 Introduction
Histopathological image analysis refers to the use of microscopical images from histological sections in order to analyze, diagnose and prevent diseases. These images are often used to obtain a high-resolution three-dimensional (3D) reconstruction of the original tissue architecture. Even with recent advances in 3D medical imaging techniques, such as Magnetic Resonance (MR) and Computed Tomography (CT), histology imaging still presents superior resolution and it remains the main source of information for several kinds of diseases, including most types of cancer [6, 8].
Several studies tackle the problem of reconstructing 3D structures from a given set of microscopic images of histological sections obtained from a single target tissue [7, 10]. These reconstructed images can be used for the study and visualization of the anatomical structures themselves, or for registration between the microscopic images and a corresponding MR macro image for multiscale analysis [4].
Given a series of images \({I_i}\left( \cdot \right) \) (\({i = 1,2, \ldots ,N}\)) scanned from stained thin sections of a chemically fixed tissue, a 3D reconstruction can be obtained by stacking up non-rigidly registered versions of such images. The registration is required due to independent translation, rotation and deformation of the histological images introduced by the process of sectioning the tissue and mouthing the sections into glass slides. The registered images \({J_i}\left( {\mathbf {y}} \right) = {I_i}\left( {{\psi ^{ - 1}} \circ {\mathbf {y}}} \right) \), obtained from the original images and the mapping \({\psi _i}\), are combined to create the full 3D reconstruction as follows:
Several registration methods have been proposed for 3D image reconstruction, which can be roughly classified into two categories: iconic (intensity-based) and geometric (landmark-based) methods [8]. In the first, images are registered by maximizing the similarities in intensity between corresponding pixels [1, 3], while in the second, images are registered by minimizing the distance between corresponding points (landmarks) on the images. This research focus on the later category due to its computational efficiency.
Let the coordinates of a landmark \(P_i^j\) detected from \({I_i}\left( {{u_1},{u_2}} \right) \) be denoted by \({\mathbf {u}}_i^j\), where \(j = 1,2, \ldots M\). Given two images, \({I_i}\left( {\mathbf {u}} \right) \) and \({I_{i + 1}}\left( {\mathbf {u}} \right) \), many landmark-based methods compute the mapping \({\psi _{i + 1}}\) using a criterion for evaluating the degree of match between corresponding landmark locations, e.g. \({{{\left\| {{\mathbf {u}}_i^j - {\psi _{i + 1}} \circ {\mathbf {u}}_{i + 1}^j} \right\| }^2}}\) [2, 11]. In other words, these methods prefer 3D images in which the corresponding landmarks are vertically aligned parallel to the \(y_3\) axis. However, corresponding landmarks are often detected along an anatomical structure which is not necessarily all vertical. The criterion employed by these methods is inconsistent with 3D microscope images, and thus often results in unnatural large deformation. In contrast, a few registration methods use a criterion that evaluates not the location matching, but the smoothness of landmark trajectories [5], which is more consistent with real tissue architecture and hence is the criterion adopted by the proposed method.
The proposed method detects corresponding landmarks using template matching, and rejects unreliable landmarks based on its confidence. As a result, the trajectories of landmarks will be automatically terminated, for instance, at blurred or folded portions, which should not contain landmarks. Once the corresponding landmarks are detected from all given images, the non-rigid mapping of each image is simultaneously determined based on the smoothed trajectories. This strategy for automatically handling damaged image portions and the capability of processing large sets of images with multiple stains, together with the results of the KPC mouse pancreas reconstruction, are the key contributions of this research.
2 Proposed Method
The reconstruction is performed over a series of N microscopic images \({I_i}\left( {\mathbf {u}} \right) \) acquired from a single tissue, where \(i = 1,2, \ldots N\) and \({\mathbf {u}} = {\left( {{u_1},{u_2}} \right) ^T}\) corresponds to coordinates on the original images. The process starts by roughly aligning the images by rigid registration [7], creating a new set \({{\tilde{I}}_i}\left( {\mathbf {x}} \right) = {I_i}\left( {{\rho ^{ - 1}} \circ {\mathbf {u}}} \right) \), where \({\rho _i}\) is the rigid transformation and \({\mathbf {x}} = {\rho _i}\left( {\mathbf {u}} \right) \).
The method then detects a set of corresponding landmark points \(P_i^j\) along the series of images \({{\tilde{I}}_i}\left( {\mathbf {x}} \right) \). Another set of destination coordinates for the landmarks must then be calculated in order to correctly deform the images. Assuming that a set of corresponding points are usually detected along several cross-sections of the same anatomical structure, and that these structures are spatially smooth and continuous, the objective is to deform the image in order that each set of corresponding points is located along a smooth curve on the reconstructed image.
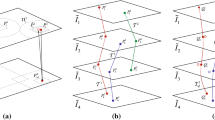
Corresponding landmarks \(P_i^j\) and trajectories \({\mathcal {T}^j}\) (center), detected from four input images \({{\tilde{I}}_i}\left( \cdot \right) \) (left). Each trajectory \({\mathcal {T}^j}\) spans between \({s^j}\) and \({t^j}\), and the set of indexes of trajectories intersecting each plane \({x_3} = i\) is given by \({{\mathcal {J}_i}}\) (right).
The process is illustrated in Fig. 1. Let \({\mathcal {P}^j} = \left\{ {P_i^j\left| {i \in \left[ {{s^j},{t^j}} \right] } \right. } \right\} \) denote the \(j^{th}\) set of corresponding landmarks detected from a subset of N images, where \(1 \leqslant {s^j} < {t^j} \leqslant N\), i.e. a set of corresponding points might be detected along only some of the N available images. Let \(\tilde{R}\left( {{x_1},{x_2},{x_3}} \right) = {{\tilde{I}}_i}\left( {{x_1},{x_2}} \right) \) denote a starting reconstruction obtained by stacking \({{\tilde{I}}_i}\left( {\mathbf {x}} \right) \), where \({x_3} = i\). The corresponding landmarks in \({\mathcal {P}^j}\) will then form a polygonal trajectory \({\mathcal {T}^j} = P_{{s^j}}^jP_{{s^{j + 1}}}^j \ldots P_{{t^j}}^j\) in \(\tilde{R}\left( {{x_1},{x_2},{x_3}} \right) \) that extends from \({x_3} = {s^j}\) to \({x_3} = {t^j}\). Due to the independent non-rigid deformations of the histological images, these trajectories usually present a non-smooth jagged shape.
The destination coordinates of each landmark point are obtained by smoothing each trajectory \({\mathcal {T}^j}\). The corresponding smoothed trajectory is denoted by \(\mathcal {T}_\mathcal {Q}^j = Q_{{s^j}}^jQ_{{s^{j + 1}}}^j \ldots Q_{{t^j}}^j\), where \(Q_i^j\) is the intersection between the smoothed trajectory with the plane \({x_3} = i\). The set \(\left\{ {Q_i^j\left| {j \in {\mathcal {J}_i}} \right. } \right\} \) of all intersections crossing this plane, where \({\mathcal {J}_i}\) is the set of indexes of trajectories crossing the plane \({x_3} = i\), is used to define a diffeomorphic mapping \({\phi _i}\) of \({{\tilde{I}}_i}\left( \cdot \right) \). The image \({{\tilde{I}}_i}\left( {\mathbf {x}} \right) \) is then non-rigidly deformed with the obtained mapping in order to transport \(P_i^j\) to \(Q_i^j\left( {j \in {\mathcal {J}_i}} \right) \). The proposed method then reconstructs a 3D image \(R\left( {{y_1},{y_2},{y_3}} \right) = {J_{{y_3}}}\left( {{y_1},{y_2}} \right) = {{\tilde{I}}_i}\left( {\phi _i^{ - 1} \circ {\mathbf {y}}} \right) \), where \({y_3} = i\), \({\mathbf {y}} = {\left( {{y_1},{y_2}} \right) ^T}\), \({\mathbf {y}}_i^j = {\phi _i} \circ {\mathbf {x}}_i^j\) and \({\mathbf {x}}_i^j\) and \({\mathbf {y}}_i^j\) are, respectively, the coordinates of \(P_i^j\) and \(Q_i^j\) in the image \({{\tilde{I}}_i}\left( {\mathbf {x}} \right) \).
Thus, the coordinates of the new points \(Q_i^j\) are obtained from \({t^j} - {s^j} + 1\) images \({{\tilde{I}}_i}\left( {i = {s^j}, \ldots ,{t^j}} \right) \) simultaneously. It is worth mentioning that similar smooth trajectories for the landmarks cannot always be obtained by simply introducing rigidity regularization to the image deformation.
In order to obtain an accurate and stable detection of corresponding points, a coarse-to-fine approach is employed. The template size D is reduced as \(D \leftarrow \gamma D\) with \(0< \gamma < 1\) after all the images \({{\tilde{I}}_i}\left( {i = 1,2, \ldots ,N} \right) \) are deformed by the mapping \({\phi _i}\). The locations of corresponding landmarks are then updated and new trajectories are obtained, and the whole process is repeated.
2.1 Template Matching
Different techniques can be employed in the detection of corresponding landmarks, such as the Scale-Invariant Feature Transform (SIFT) or the Normalized Cross-Correlation (NCC), the latter being the method used in this research. Initially, a set of points \(P_i^j\left( {j = 1,2, \ldots } \right) \) is iteratively sampled from \({{\tilde{I}}_i}\left( \cdot \right) \) with probability \(p_i^j\) proportional to \(\left\| {\nabla {{\tilde{I}}_i}\left( {\mathbf {x}} \right) } \right\| \). New landmarks are only sampled from coordinates at least D pixels away from any existing landmarks.
False matchings are suppressed based on the confidence of the template matching given by the NCC and by applying backward template matching [13]. First, any landmark with an NCC value lower than a threshold is eliminated. After, backward template matching is applied in order to detect a landmark \(\hat{P}_i^j\) corresponding to \(\tilde{P}_{i + 1}^j\). The landmark \(P_{i + 1}^j\) is discarded if the distance between \(P_i^j\) and \(\hat{P}_i^j\) is larger than a threshold.
2.2 Image Warping
The proposed method smooths the trajectories \({\mathcal {T}^j}\) in order to obtain the destination points \(Q_i^j\). The coordinates \({\mathbf {y}}_i^j\) of the destination points are calculated by minimizing the square of the total variation of each trajectory and their square errors with a tradeoff parameter \(\lambda \) as follows:
This function is convex and has a unique minimizer, which can be analytically calculated. With the obtained destination coordinates \({\mathbf {y}}_i^j\) for each landmark \({\mathbf {x}}_i^j\), the diffeomorphic mapping \({\phi _i}\) can be computed using the well-known B-spline deformation method [9], which allows a diffeomorphic mapping with hard constrains \({\mathbf {y}}_i^j = {\phi _i}\left( {{\mathbf {x}}_i^j} \right) \).
3 Experimental Results
The reconstruction method was evaluated using a dataset of histological sections from the pancreas of a KPC mouse [12]. The extracted tumorous pancreas was fixed with formaldehyde, split into five 5 mm blocks, and each was sliced in a series of 4 \(\upmu \)m-thick sections. After damaged samples were removed, the dataset contains around 2500 images, the majority (85%) stained with Hematoxylin & Eosin (HE) stain, but some with Antigen KI-67 (Ki67), Masson’s Trichrome (MT) and Cytokeratin-19 (CK19) stains, which are interposed between HE stained images. The size of the original images is 100k \(\times \) 60k pixels; however, a downsampled version of 15k \(\times \) 10k pixels was used in some of the experiments.
Figure 2 shows actual calculated trajectories from a portion of the images. The original trajectories \({\mathcal {T}^j}\) are shown in Fig. 2(a), while the smoothed trajectories \(\mathcal {T}_\mathcal {Q}^j\) are shown in Fig. 2(b). Figure 3 shows cross-sections of \(\tilde{R}\left( \cdot \right) \) and \(R\left( \cdot \right) \), i.e. before and after smoothing. In Fig. 3(a), even the major structures are difficult to discern, while small structures, such as blood vessels and ducts, are virtually indistinguishable. The smoothed version shown in Fig. 3(b) enables the observation of fine details and small structures, which can then be labeled for further analysis. The hyperparameters were determined experimentally and are not critical for the overall performance of the method.
Visualization of the internal microstructures from a portion of the full 3D reconstruction of a block of images: (a) registered HE stained image portion, (b) labeled necrosed portion of the tumor, (c) labeled pancreatic ducts, and (d) combined 3D reconstruction of ducts (blue and green) and necrosis (red). (Color figure online)
Labeling, detection and reconstruction of active regions within the tumor from Ki67 stained images: (a) Ki67 stained image portion, (b) automatic labeling of active cells (brown spots), (c) high density (red) regions of active cells (overall image), and (d) reconstruction of active areas and superposition with full block 3D reconstruction, which contains 810 images of 15k \(\times \) 10k pixels, or around \(121.5 \times {10^9}\) voxels. (Color figure online)
It is then possible to automatically label and reconstruct the structure of the central necrosed portion of the tumor and the pancreatic ducts, as shown in Fig. 4. This reconstruction reveals a high concentration of ducts around the necrosis.
Different microstructures require different stains in order to be accurately identified and labeled. Figure 5 shows registration results for neighbor sections of different stains, confirming the robustness of the proposed registration method. The Ki67 stain can indicate high cell proliferation. After reconstruction, sections stained with Ki67 were automatic labeled based on hue and the density of brown spots was calculated using Kernel Density Estimation (KDE), as shown in Fig. 6. By linearly interpolating along all sections and combining this results with the anatomical reconstruction of the tumor block, it is possible to observe the high proliferation regions on the external portion of the tumor.
4 Conclusions
This paper describes a non-rigid registration method for 3D reconstruction from microscopic images of histological sections. The method explicitly constructs smooth trajectories in order to determine the deformation of all images simultaneously. This approach suppresses the unnatural vertical structures often created by conventional landmark-based methods, which process images pairwise.
Experimental results confirm the efficiency of the registration procedure, using a large dataset of histological images with different stains from the pancreas of a KPC mouse. The obtained reconstructions present continuous and smooth anatomical micro-structures, which could be successfully labeled and visualized.
Future work will include the registration of a new full-body KPC mouse image dataset, as well as a quantitative analysis of the registration results.
References
Beg, M.F., Miller, M.I., Trouvé, A., Younes, L.: Computing large deformation metric mappings via geodesic flows of diffeomorphisms. Int. J. Comput. Vis. 61(2), 139–157 (2005)
Beg, M.F., Miller, M., Trouvé, A., Younes, L.: The euler-lagrange equation for interpolating sequence of landmark datasets. In: Ellis, R.E., Peters, T.M. (eds.) MICCAI 2003. LNCS, vol. 2879, pp. 918–925. Springer, Heidelberg (2003). https://doi.org/10.1007/978-3-540-39903-2_112
Cifor, A., Bai, L., Pitiot, A.: Smoothness-guided 3-D reconstruction of 2-D histological images. NeuroImage 56(1), 197–211 (2011)
Dauguet, J., et al.: Three-dimensional reconstruction of stained histological slices and 3D non-linear registration with in-vivo MRI for whole baboon brain. J. Neurosci. Methods 164(1), 191–204 (2007)
Gaffling, S., Daum, V., Hornegger, J.: Landmark-constrained 3-D histological imaging: a morphology-preserving approach. In: Vision, Modeling, and Visualization, pp. 309–316 (2011)
Gurcan, M.N., Boucheron, L.E., Can, A., Madabhushi, A., Rajpoot, N.M., Yener, B.: Histopathological image analysis: a review. IEEE Rev. Biomed. Eng. 2, 147–171 (2009)
Ourselin, S., Roche, A., Subsol, G., Pennec, X., Ayache, N.: Reconstructing a 3D structure from serial histological sections. Image Vis. Comput. 19(1), 25–31 (2001)
Pichat, J., Iglesias, J.E., Yousry, T., Ourselin, S., Modat, M.: A survey of methods for 3D histology reconstruction. Med. Image Anal. 46, 73–105 (2018)
Rueckert, D., Aljabar, P., Heckemann, R.A., Hajnal, J.V., Hammers, A.: Diffeomorphic registration using B-splines. In: Larsen, R., Nielsen, M., Sporring, J. (eds.) MICCAI 2006. LNCS, vol. 4191, pp. 702–709. Springer, Heidelberg (2006). https://doi.org/10.1007/11866763_86
Saalfeld, S., Cardona, A., Hartenstein, V., Tomančák, P.: As-rigid-as-possible mosaicking and serial section registration of large ssTEM datasets. Bioinformatics 26(12), i57–i63 (2010)
Sotiras, A., Davatzikos, C., Paragios, N.: Deformable medical image registration: a survey. IEEE Trans. Med. Imaging 32(7), 1153–1190 (2013)
Westphalen, C.B., Olive, K.P.: Genetically engineered mouse models of pancreatic cancer. Cancer J. 18(6), 502–510 (2012)
Yin, Z., Collins, R.: Moving object localization in thermal imagery by forward-backward MHI. In: Proceedings of the Computer Vision and Pattern Recognition Workshop, pp. 133–133. IEEE (2006)
Acknowledgments
This research was supported by JSPS KAKENHI Grant Number 26108003.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Kugler, M. et al. (2018). Accurate 3D Reconstruction of a Whole Pancreatic Cancer Tumor from Pathology Images with Different Stains. In: Stoyanov, D., et al. Computational Pathology and Ophthalmic Medical Image Analysis. OMIA COMPAY 2018 2018. Lecture Notes in Computer Science(), vol 11039. Springer, Cham. https://doi.org/10.1007/978-3-030-00949-6_5
Download citation
DOI: https://doi.org/10.1007/978-3-030-00949-6_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00948-9
Online ISBN: 978-3-030-00949-6
eBook Packages: Computer ScienceComputer Science (R0)