Abstract
Brain tumor localization and segmentation is an important step in the treatment of brain tumor patients. It is the base for later clinical steps, e.g., a possible resection of the tumor. Hence, an automatic segmentation algorithm would be preferable, as it does not suffer from inter-rater variability. On top, results could be available immediately after the brain imaging procedure. Using this automatic tumor segmentation, it could also be possible to predict the survival of patients. The BraTS 2018 challenge consists of these two tasks: tumor segmentation in 3D-MRI images of brain tumor patients and survival prediction based on these images. For the tumor segmentation, we utilize a two-step approach: First, the tumor is located using a 3D U-net. Second, another 3D U-net – more complex, but with a smaller output size – detects subtle differences in the tumor volume, i.e., it segments the located tumor into tumor core, enhanced tumor, and peritumoral edema.
The survival prediction of the patients is done with a rather simple, yet accurate algorithm which outperformed other tested approaches on the train set when thoroughly cross-validated. This finding is consistent with our performance on the test set - we achieved 3rd place in the survival prediction task of the BraTS Challenge 2018.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Brain tumors can appear in different forms, shapes and sizes and can grow to a considerable size until they are discovered. They can be distinguished into glioblastoma (GBM/HGG) and low grade glioma (LGG). A common way of screening for brain tumors is with MRI-scans, where even different brain tumor regions can be determined. In effect, MRI scans of the brain are not only the basis for tumor screening, but are even utilized for pre-operative planning. Thus, an accurate, fast and reproducible segmentation of brain tumors in MRI scans is needed for several clinical applications.
HGG patients have a poor survival prognosis, as metastases often develop even when the initial tumor was completely resected. Whether patient overall survival can be accurately predicted based on pre-operative scans by employing knowing factors such as radiomics features, tumor location and tumor shape, remains an open question.
The BraTS challenge [11] addresses these problems, and is one of the biggest and well known machine learning challenges in the field of medical imaging. Last year around 50 different competitors from around the world took part. This year, the challenge is divided in two parts: First, tumor segmentation based on 3D-MRI images, and second, survival prediction of the brain tumor patients based on only the pre-operative scans and the age of the patients.
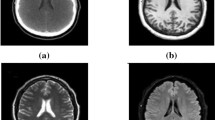
Example of image modalities and groundtruth-labels in the BraTS 2018 dataset. The subtraction image is calculated by subtracting the T1 image (a) from the T1 post-contrast image (b), as described in Sect. 3.1. For the labels, blue indicates the peritumoral edema, green the necrotic and non-enhancing tumor, and red the GD-enhancing core, as described in the BraTS paper [11]. (Color figure online)
Similar to the BraTS 2017 dataset, the BraTS 2018 training dataset consists of MRI-scans of 285 brain tumor patients from 19 different contributors. The dataset includes T1, T1 post-contrast (T1CE), T2, and T2 Fluid Attenuated Inversion Recovery (Flair) volumes, as well as hand-annotated expert labels for each patient [1,2,3]. An example of a set of images can be seen in Fig. 1.
Motivated by the success of the U-net [14] in biomedical image segmentation, we choose the 3D-adaptation [5] of this architecture to tackle the segmentation part of the BraTS challenge. Two different versions are used, a first one to coarsely locate the tumor, and a second one to accurately segment the located tumor into different areas.
Concerning the survival prediction, we found that complex models using different types of radiomics features such as shape and texture of the tumor and the brain could not outperform a simple linear regressor based on just a few basic features. Using only the patient age and tumor region sizes as features, we achieve competitive results.
The code developed for this challenge is available online: https://github.com/weningerleon/BraTS2018.
2 Related Work
In the last years, deep learning has advanced classification and segmentation in many biomedical imaging applications, and has a preeminent role in current publications.
In the BraTS Challenge last year, all top-ranking approaches of the segmentation task [6, 9, 16, 17] used deep convolutional neural networks. The employed architectures vary substantially among these submission. However, a common ground seems to be the utilization of 3D-architectures instead of 2D-architectures.
One key architecture for biomedical segmentation, which is also heavily used throughout this paper, is the U-Net [14]. Both, 2D as well as 3D-variants [5] have been successfully employed for various biomedical applications, and still achieve competitive results in current biomedical image segmentation challenges [7, 8].
3 Methods
3.1 Segmentation
We tackle the segmentation task in a two-step approach: First, the location of the brain tumor is determined. Second, this region is segmented into the three different classes: peritumoral edema (ed), necrotic tumor (nec), and GD-enhancing core (gde).
Preprocessing. We first define a brain mask based on all voxels unequal to zero, on which all preprocessing is carried out. On this brain mask, the mean and standard deviation of the intensity is calculated, and the data normalized accordingly. Since different MRI-scanners and sequences are used, we independently normalize each image and modality based on the obtained values. Non-brain regions remain zero.
The whole tumor is strongly visible in T1, T2 and Flair MRI-images. However, in practice, including all images seems to produce better results even for the whole tumor localization. We also add another image as input, a contrast-agent subtraction image, where the T1 image is subtracted from the T1CE image. This should enhance the contrast-agent sensitive region, as can be seen in Fig. 1c.
We construct a cuboid bounding box around the brain, and crop the non-brain regions to facilitate training. The training target is constructed by merging the three different tumor classes of the groundtruth labels.
For training of the tumor segmentation step, the 3D-images are cropped around a padded tumor area, which is defined as the area of 20 voxels in every direction around the tumor.
Network Architectures and Employed Hardware. For both steps, a 3D U-net [5] with a depth of 4 is employed.
The first U-net uses padding in every convolutional layer, such that the input size corresponds exactly to the output size. Every convolutional layer is followed by a ReLU activation function. 16 feature maps are used in the first layer, and the number of feature maps doubles as the depth increases. For normalization between the different layers, instance-norm layers [15] are used, as they seem to be better suited for normalization in segmentation tasks and for small batch sizes. Testing different training hyperparameters, the Adam optimizer [10] with an initial learning rate of 0.001 together with a binary cross entropy loss was chosen for the tumor localization step. An L2-regularization of 1e−5 is applied to the weights, and the learning rate was reduced by a factor of 0.015 after every epoch. One epoch denotes a training step over every brain.
The U-net utilized in the second step has a similar architecture as the previous one, but with double as many feature maps per layer. To counteract the increased memory usage, no padding is used, which drastically reduces the size of the output as well as the memory consumption of later feature maps.
Here, we apply a multi-class dice loss to the output of our 3D U-net and the labels for training, as described in [12]. A learning rate of 0.005 was chosen, while weight decay and learning rate reduction remain the same as in step 1.
Our contribution to the BraTS challenge was implemented using pyTorch [13]. Training and prediction is carried out on a Nvidia 1080 Ti GPU with a memory size of 11 Gb.
Training. In the first step, we train with complete brain images cropped to the brain mask. The brain mask is determined by all voxels not equal to zero. Using a rather simple U-net, a training pass with a batch-size of one fits on a GPU even for larger brains. Due to the bounding box around the brain, different sizes need to be passed through the network. In practice this is possible using a fully convolutional network architecture and a batch size of one.
For the second step, we choose the input to be fixed to \(124\times 124\times 124\). Due to the unpadded convolutions, this results in an output shape of \(36\times 36\times 36\). Hence, the training labels are the \(36\times 36\times 36\) sized segmented voxels in the middle of the input. Here, a batch-size of two was chosen.
During training, patches are chosen from inside the padded tumor bounding box for each patient. To guarantee a reasonably balanced train set, only training patches which comprise all three tumor classes are kept for training.
Inference. Similar to the training procedure, the first step is carried out directly on a complete 5-channel (T1, T2, Flair, T1CE, and contrast-agent subtraction image) 3D image of the brain.
Before the tumor/non-tumor segmentation of this step is used as basis in the second step, only the largest connected area is kept. Based on the assumption that there is only one tumorous area in the brain, we can suppress false positive voxels in the rest of the brain with this method.
We then predict \(36\times 36\times 36\) sized patches with the trained unpadded U-net. Patches are chosen so that they cover the tumorous area, and the distance between two neighboring patches was set to 9 in each direction. This results in several predictions per voxel. Finally, a majority vote over these predictions gives the end result.
3.2 Survival Prediction
According to the information given by the segmentation labels, we count the number of voxels of the tumor segmentation. This volume information about the necrotic tumor core, the GD-enhancing tumor and peritumoral edema as well as the distance between the centroids of tumor and brain and the age of the patient were considered as valuable feature for the survival prediction task. We tested single features, as well as combinations of features as input for a linear regressor.
4 Results
4.1 Segmentation
For evaluation on the training dataset, we split the training dataset randomly into 245 training images and 40 test images to evaluate our approach with groundtruth labels. No external data was used for training or pre-training.
Based on our experience with the training dataset, we choose 200 epochs as an appropriate training duration for the first step, and 60 epochs as an appropriate training duration for the second step. We thus train from scratch on all training images for the determined optimal number of epochs, and use the obtained networks for evaluation on the validation set. The results obtained by this method can be seen in Table 1, and an exemplary result is visualized in Fig. 2.
4.2 Survival Prediction
For evaluating our approach on the training dataset, we fit and evaluate our linear regressor with a leave-one-out cross-validation on the training images. We compare the results obtained by solely using the age of the patient versus using the age with a subset of the tumor region sizes as features. On top, we consider the distance between the centroid of the tumor and the centroid of the brain as a feature. Our finding is that all features other than the age of the patient increase the error on left-out images. In Tables 2 and 3, we show the exact results for the different input features on the training set (cross-validation) and on the test set (according to the online portal).
In Fig. 3, the survival time in years is plotted against the age for all patients with a resection status of ‘gross total resection’ in the train dataset. The linear regressor fitted to this data and used for the challenge, is plotted as well. The three classes used during the challenge, dividing the dataset into long, short, and mid-survivors can also be seen.
This age-only linear regressor achieved the 3rd place in the BraTS challenge 2018 [4], with an accuracy of 0.558, a MSE of 277890 and a median SE of 43264 on the test data.
5 Discussion and Conclusion
Our contribution submitted to the BraTS challenge 2018 was summarized in this paper. We used a two-step approach for tumor segmentation and a linear regression for survival prediction.
The segmentation approach already gives promising results. In practice, the two-step framework helps eliminating spurious false-positive classifications in non-tumorous areas, as only the largest connected area is considered as tumor. However, this assumes that there is only one tumorous area in the brain. As there is only one tumorous area in the vast majority of cases, this boosts the accuracy measured. Notwithstanding, it is a simplification that can lead to serious misclassifications in single cases.
This simplification needs to be tackled in future development of the framework. Furthermore, we will evaluate a broader variety of different network architectures, and will also include 3D data-augmentation techniques into our framework.
Our algorithm for the survival analysis task is a straight-forward approach. We considered other, more complex approaches, which were however not able to beat this baseline algorithm.
On the validation set, our survival prediction algorithm ranks among the top submissions, e.g., the age-only approach achieves the lowest MSE and second highest accuracy according to the online portal.
Finally, our top-placement (3rd place) in the challenge underlines the strength of the age as feature for survival prediction. Other teams, using various radiomics and/or deep learning approaches, could not perform much better than our straight-forward approach. Hence, it can be concluded that pre-operative scans are not well suited for survival prediction. However, other datasets could be better suited for survival prediction, e.g., post-operative or follow-up scans of the patient.
References
Bakas, S., et al.: Segmentation labels and radiomic features for the pre-operative scans of the TCGA-LGG collection. The Cancer Imaging Archive (2017)
Bakas, S., et al.: Segmentation labels and radiomic features for the pre-operative scans of the TCGA-GBM collection. The Cancer Imaging Archive (2017)
Bakas, S., et al.: Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Scientific Data 4, 170117 EP, September 2017. https://doi.org/10.1038/sdata.2017.117
Bakas, S., et al.: Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv preprint arXiv:1811.02629
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O.: 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 424–432. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_49
Isensee, F., Kickingereder, P., Wick, W., Bendszus, M., Maier-Hein, K.H.: Brain tumor segmentation and radiomics survival prediction: contribution to the BRATS 2017 challenge. CoRR abs/1802.10508 (2018). http://arxiv.org/abs/1802.10508
Isensee, F., Kickingereder, P., Wick, W., Bendszus, M., Maier-Hein, K.H.: No new-net. CoRR abs/1809.10483 (2018). http://arxiv.org/abs/1809.10483
Isensee, F., et al.: nnU-Net: self-adapting framework for U-Net-based medical image segmentation. CoRR abs/1809.10486 (2018). http://arxiv.org/abs/1809.10486
Kamnitsas, K., et al.: Ensembles of multiple models and architectures for robust brain tumour segmentation. In: Crimi, et al. [6], pp. 450–462. https://doi.org/10.1007/978-3-319-75238-9_38
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. CoRR abs/1412.6980 (2014). http://arxiv.org/abs/1412.6980
Menze, B.H., et al.: The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imag. 34(10), 1993–2024 (2015). https://doi.org/10.1109/TMI.2014.2377694
Milletari, F., Navab, N., Ahmadi, S.: V-Net: Fully convolutional neural networks for volumetric medical image segmentation. CoRR abs/1606.04797 (2016). http://arxiv.org/abs/1606.04797
Paszke, A., et al.: Automatic differentiation in PyTorch. In: NIPS-W (2017)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015, part III. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Ulyanov, D., Vedaldi, A., Lempitsky, V.S.: Instance normalization: the missing ingredient for fast stylization. CoRR abs/1607.08022 (2016). http://arxiv.org/abs/1607.08022
Wang, G., Li, W., Ourselin, S., Vercauteren, T.: Automatic brain tumor segmentation using cascaded anisotropic convolutional neural networks. In: Crimi, et al. [6], pp. 178–190. https://doi.org/10.1007/978-3-319-75238-9_16
Yang, T.L., Ou, Y.N., Huang, T.Y.: Automatic segmentation of brain tumor from MR images using SegNet: selection of training data sets. In: Crimi, et al. [6], pp. 450–462. https://doi.org/10.1007/978-3-319-75238-9
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Weninger, L., Rippel, O., Koppers, S., Merhof, D. (2019). Segmentation of Brain Tumors and Patient Survival Prediction: Methods for the BraTS 2018 Challenge. In: Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., van Walsum, T. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2018. Lecture Notes in Computer Science(), vol 11384. Springer, Cham. https://doi.org/10.1007/978-3-030-11726-9_1
Download citation
DOI: https://doi.org/10.1007/978-3-030-11726-9_1
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-11725-2
Online ISBN: 978-3-030-11726-9
eBook Packages: Computer ScienceComputer Science (R0)