Abstract
We describe a method to automatically predict scoliosis in Dual-energy X-ray Absorptiometry (DXA) scans. We also show that intermediate representations, which in our case are segments of body parts, help improve performance. Hence, we propose a two step process for prediction: (i) we learn to segment body parts via a segmentation Convolutional Neural Network (CNN), which we show outperforms the noisy labels it was trained on, and (ii) we predict with a classification CNN that uses as input both the raw DXA scan and also the intermediate representation, i.e. the segmented body parts. We demonstrate that this two step process can predict scoliosis with high accuracy, and can also localize the spinal curves (i.e. geometry) without additional supervision. Furthermore, we also propose a soft score of scoliosis based on the classification CNN which correlates to the severity of scoliosis.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
Scoliosis is an abnormal sideways curvature of the spine typically occuring prior to puberty and affects approximately 1.1% to 2.9% of children [12]. While most cases are mild, stablizing over time and presenting few symptoms, some children develop severe deformaties that can cause lifelong disability and pain. Scoliosis can also cause back pain [1] and in rare cases can cause respiratory failure [8]. It is not currently possible to determine prognosis at the onset of disease and hence children with scoliosis are monitored with repeated X-Ray imaging to determine whether the disease is stable or progressing. While accepted as the standard of care, the use of repeated X-Ray imaging on children with the associated radiation dose is far from ideal. Moreover, the radiation dose also precludes its use in population based epidemiological studies to better understand disease progression and develop future tools to predict prognosis and for screening.
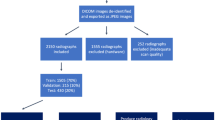
DXA Scans: The use of DXA imaging for diagnosis and monitoring of scoliosis has been proposed as an alternative to X-Ray due to its very low radiation dose compared to spinal X-Rays (0.001 mSv vs. 1.5 mSv) and widespread availability [12]. DXA scans, typically used to measure bone mineral density in the management of osteoporosis, are whole body scans acquired in a line scanning manner from the top of the head to the bottom of the feet. Two X-Ray sources at different energy levels are used to create a pair of absorption images which are then post-processed to produce quantitative bone mineral density images. While detection of scoliosis using DXA has been shown to be feasible and accurate, the manual technique proposed by [12] is labour intensive and requires careful adherence to the prescribed analysis protocol for accurate results. That being said, the method has proven to be quite successful in scoliosis research e.g. [5]. The technique involves first localizing important body parts to establish a reference coordinate system. These are then used for two purposes: (i) the head and legs are used to determine the overall body position because incorrect positioning can either mask or mimic the appearance of the condition, and (ii) the curvature of the spine is used to assess for the presence of the condition; defined to be when the curvature is  . Our goal in this work is to automate the process of scoliosis classification using DXA, based on [12]. An overview of our approach is given in Fig. 1.
. Our goal in this work is to automate the process of scoliosis classification using DXA, based on [12]. An overview of our approach is given in Fig. 1.
Intermediate Representations: Our approach is based on a CNN driven by a set of intermediate representations that attempt to mimic the intuition of the underlying process of [12] described above. Our hypothesis is that such intermediate representations, in this case soft-segmentation masks, can improve classification performance, at least within the context of specific medical applications and when training with dataset sizes typically available in medical image analysis. The intermediate representations embed prior knowledge on how scoliosis is imaged and assessed in the case of DXA, and provide important cues for the network. In more detail, we provide several soft map segmentations of the key parts of the anatomy used in the DXA assessment process: the head and legs, to determine the overall body position; and the spine, so that its curvature can be used to assess for the presence and severity of the condition. In effect, the use of such intermediate representations guides the learning process to focus on important parts in determining scoliosis.
Related Work: Intermediate representations have recently been proposed as a means to extract characteristic object representations in the MarrNet 2.5D sketches by [14], and to take advantage of available training datasets for learning keypoints by [15]. Our use of intermediate representations differs from these. There has been a lot of work done on whole body DXA scans e.g. manual segmentation of body parts in [2] and modelling the shape of the body in [11]. There is also work looking at the spine using DXA, more specifically segmenting the vertebral body [9] but ours is the first system to segment the spine automatically in whole body DXA scans.
Contributions and Overview: This paper makes several contributions: (i) we present an automated method to predict scoliosis from DXA scans; (ii) we demonstrate improved classification performance of scoliosis when DXA images are augmented with application tuned intermediate representations; (iii) we illustrate how such intermediate representations may be robustly generated using a network trained on “cheaply” obtained but noisy labels; and (iv) we propose that our network can infer a continuous scale of the severity of scoliosis even though it has been trained on binary labels. The remainder of the paper is organized in two main sections: Sect. 2 describes the approach and process by which we train a (segmentation) network for generating the intermediate representations. Section 3 describes the network for predicting the scoliosis and related labels from both the DXA scans and intermediate representations. The description of the dataset and experimental results then follow in Sects. 4 and 5 respectively, including a proposal for a scoliosis score, and evidence hotspots localizing the curvature of the spine.
2 Segmentation
There are multiple body parts that can be seen in the whole body DXA scans, not all are important for predicting scoliosis. Hence, a sensible approach to automate prediction of scoliosis from these scans is to segment relevant body parts prior to classification of scoliosis. The body parts we segment are: (1) head, (2) spine, (3) pelvis, (4) pelvic cavity, (5) left leg, and (6) right leg. The spine is the most important part since scolios is a disease of the spine while the others are important for predicting positioning error (straight body vs. curved). Positioning error also plays a part in determining scoliosis as the orientation of the head and legs also affects curvature of the spine.
Since the full body DXA scans are homogeneous, segmentation labels for some parts of the body can be produced with a series of simple heuristics. These labels, although not perfect, are good enough to train a segmentation CNN and, as will be seen, in many cases the trained CNN produces visually better segmentations. In the following sections we describe the stages of training the segmentation CNN: first, generating (possibly noisy) segmentation labels using simple heuristics from classical computer vision; and second, defining loss functions and the architecture of the CNN.
2.1 Generating Segmentation Labels
For each scan, the head is first segmented via active contour around the head region [3]. The pelvis is located by scanning each row of the image starting from the bottom of the image until the bimodal intensity from the legs becomes unimodal. Working through the body in this way, using a combination of active contours and row based intensity modes, each of the body parts in turn can be segmented. Note, this is only possible because of the uniform positioning of the body adopted for the DXA scans. Around 90% of the scans are good though rough. Examples of the segmentation masks from these simple heuristics can be seen in Fig. 2.
2.2 A CNN for Segmentation
The goal is to automatically segment the labelled body parts for each DXA scan using a CNN. The segmentation CNN takes in a DXA scan as input and produces six different channels with same dimension as the input, where each channel corresponds to the six labelled parts as shown in Fig. 2. The design of the network is inspired by the U-Net architecture with minor changes [10]. The architecture of the network is given in Fig. 3.
Segmentation CNN: the network takes in a full body DXA scan and produces segmentation masks for each of the six body parts. The network is based on the U-Net architecture in that we have multiple skip connections from the earlier layers of the network connecting to the later layers. / 2 denotes a stride of 2. The output shows the segmentation output overlaid on top of the input (in actuality the network only produces the segmentation mask).
Segmentation Losses: We consider two different losses to train the segmentation network. The first is a standard \(L_2\) loss:

where \(y_n\) is segmentation label (binary, \(y = 1\) for parts containing a body part and \(y = 0\) otherwise), and \(\hat{y}_n\) is the output of the network for sample n. The loss is also balanced by the amount of background and foreground pixels in the batch during training.
Inspired by the method of which DXA scanners typically operate (similar to a line scan camera); scan line by scan line or row by row of the whole scan, we also propose a segmentation loss on a per scan line basis. This is done as follows: for each scan line, the network is tasked to predict both the mid-point and thickness of the labelled body part. The mid-point prediction can be viewed as a 128-way classification task where each class is the point of the 128-dimensional scan line (i.e. the width of the image), optimized via a standard softmax log loss:
where \(y_j\) is the jth component of the Conv11 output for \(x_n\) per scan line. The raw output of this layer is a mid-point heatmap for each labelled body part. The prediction of thickness for each scan line can be expressed as the summation of the number of pixels belonging to a labelled body part e.g. a scan line with 8 spine pixels would have a thickness of 8 for the spine class. The same Conv11 output \(y_j\) is used for predicting the thickness, optimized with \(L_2\) loss:

where \(\mathcal {H}\) is the Heaviside step function which is approximated via a sigmoid, used to binarize the activation of the Conv11 output:
where k controls the steepness (\(k = 10\) in our case). To produce the segmentation mask for each scan, we combine the predicted mid-point (max of the activation of Conv11 for each scan line) and the thickness of a labelled body part for the corresponding scan line (see Fig. 4). A segmentation mask can also be produced directly after the Heaviside activation but we find this leads to be slightly worse segmentation performance.
The Benefits of Using a Segmentation CNN: Although we are able to produce segmentation masks via very simple heuristic and classical computer vision methods, in about 10% of cases there are erroneous segmentations especially for a really difficult body part like the spine. As the goal is to build an end-to-end system of scoliosis prediction, a CNN is much more suitable approach as it learns, despite the noisy training labels, to correctly predict the segmentation masks. Figure 5 shows examples of failure cases for the simple method against output of a CNN on the test set. A second benefit of using the CNN is that we obtain a ‘soft-segmentation mask’. As will be seen, using this as an intermediate representation improves the classification performance compared to using the hard segmentations.
Segmentation mask from mid-point and thickness: the segmentation masks from intermediate soft segmentation of the body parts, which contain mid-point information, alongside the corresponding thickness vector for each body part. We find the intermediate segmentation, or soft mask, from the raw output of Conv11 can also be used for classifying scoliosis and other tasks.
Simple heuristics vs CNN: “Noisy” is the noisy annotation generated via simple heuristics, and used to train the CNN. We see around 10% failure cases. Here we show examples of those failure cases on the test set compared to the CNN segmentation for the spine. Failures typically appears as under-segmentation of the spine around the base or the middle of the spine highlighted in the “Noisy” examples.
3 Classification
The goal is to predict three different classifications for each DXA scan: (i) a binary classification of scoliosis vs. non-scoliosis, outlined in [12], (ii) a binary classification of positioning error which is dependant on the straightness of the whole body in the DXA scan, and (iii) the number of curves of a scoliotic spine (only on cases with scoliosis). The number of curves is divided into three different classes: no curve (normal spine); one curve, i.e. a “C” shaped spine; and more than one curve, which includes the classical “S” shaped spine with two curves. The networks for classification share the first six layers, five convolutional and one fully connected layer, which branch out for each of the three classification tasks (see Fig. 6).
Classification CNN: The network is inspired by the VGG-M network in [4] with 5 convolutional layers and 3 fully connected layers but with slightly different filter sizes and number of filters. We experimented with several different input for the classification network: (1) raw DXA scan, (2) segmentation mask, (3) mid-point map, and (4) a combination of the raw DXA and either the segmentation mask, mid-point map or both.
Classification Loss: We follow the multi-task balanced loss approach discussed in [7] which can be expressed as minimizing a combination of the softmax log-losses of the three classifications:
where t corresponds to each classification and \(t \in \{1 \dots 3\}\), x is the input scan, \(C_t\) which corresponds to the number of classes in task t, \(y_j\) is the \(j^{th}\) component of the FC8 output, and c is the true class of \(x_n\). The loss for each classification is also balanced with the inverse of the frequency of the class to emphasize the contribution of the minority class e.g. only \(8\%\) of the scans have scoliosis.
4 Dataset and Training Details
The dataset is from the Avon Longitudinal Study of Parents and Children (ALSPAC) cohort that recruited pregnant women in the UK. The DXA scans of the subjects were obtained from two different time points; when the subjects were 9 and 15 years of age. This difference in acquisition period and the variation of height between different individuals results in a difference of scan heights. Figure 7 shows a comparison of scans from various individuals at different time points.
In all, there are 7645 unique subjects in the dataset, most of which have two scans, which totals to 12028 scans. The distribution of labels of the different classification tasks is given in Table 1. We use a 80:10:10 (train:test:validation) random split, on a per patient basis (about 9.6k:1.2k:1.2k scans). Two different random splits of the data are used throughout (from training to evaluation) in order to obtain standard deviations on the classification performance.
Pre-processing: The scans are normalized such that both the head and feet are roughly in the same region for all the scans regardless of age and original height of the scans. Empty spaces on top of the head and below the feet are also removed. The scans are cropped isotropically to prevent distortion and to keep the aspect ratio the same as the original. The dimensions of the scans after normalization is \(416 \times 128\) pixels.
Training Details: Both the segmentation and classification networks are optimized via stochastic gradient descent (SGD) from scratch. The hyperparameters are; batch size 64 for segmentation and 256 for classification; momentum 0.9; weight decay 0.0005; initial learning rate of 0.0001 for segmentation and 0.001 for classification, both of which are lowered by a factor of 10 as the loss plateaus. The network were trained via the MatConvNet [13] toolbox using an NVIDIA Titan X GPU. We employ several training augmentation strategies: (i) translation of \(\pm 24\) pixels in the x-axis, (ii) translation of \(\pm 24\) pixels in the y-axis, and (iii) random flipping. At test time, the final prediction is calculated from the average prediction of an image and its flip.
5 Experiments and Results
5.1 Segmentation
Segmentation Losses Comparison: The segmentations are evaluated using the intersection over union (IoU) between the predicted output and the noisy label generated in Sect. 2.1. A CNN is trained for each loss, and their performance compared in Table 2. The performance of the network trained on the mid-point and thickness losses outperforms the network trained on the \(L_2\) loss on every body part segmentation apart from the pelvis; 0.77 vs. 0.72. This might be due to the fact that the pelvis is a much more complex segmentation task and harder to segment on a per scan line basis. The pelvis ground truth annotations made by the simple heuristics segmentation are also a lot noisier than the other body parts.
5.2 Classification
Comparison of Input for Classification. We investigate different inputs for the CNN for predicting the three classification tasks. The different inputs are combinations of: (i) the raw DXA scan, (ii) the segmentation masks of the body parts, and (iii) a soft segmentation of the body parts obtained from the output of the Conv11 layer from the segmentation CNN (which also has mid-point information of each body part per scan line). The network which only use the raw DXA input is considered as baseline. CNNs with multiple inputs have concatenation layers after FC6 and share the last two layers for each task. The average per-class accuracy is given in Table 3. It can be seen that the best choices are networks that take in raw DXA together with either of the two intermediate representations, both hard and soft segmentation masks. Looking at each task individually, the best network for scoliosis is the CNN (E) that takes in both the raw DXA scan and the soft mask of the body parts, with an improvement of +3.8% (86.7% \(\rightarrow \) 90.5%) compared to the baseline CNN (A) that inputs just the raw DXA. CNN (E) outperforms the baseline by +3.6% (69.0% \(\rightarrow \) 72.6%) for predicting the number of curves. Finally, the best result for positioning error is CNN (D) which is +0.2% better than CNN (A) (81.5% \(\rightarrow \) 81.7%). To summarize, looking at Table 3, adding intermediate representation as input to the classification CNN is always better, and that when comparing intermediate representation, soft segmentation masks are better than hard (binary) segmentation masks.
Severity of scoliosis: shown are examples on the test set and their soft scores for scoliosis prediction; scans with scores approaching 1 are more scoliotic and scores approaching 0 are normal. In this example, the 3 examples on the left are normal scans and the 3 examples on the right have scoliosis.
Classification Hotspots. We investigate the weak localization of the task learned by the CNN or evidence hotspots as in [6, 7]. We follow the method outlined in [16]. The best task to look at in our case is the scoliosis prediction. Figure 8 shows different examples of scans in the test with scoliosis alongside their hotspots. As expected, the hotspots manage to localize the spines in the images, but also, interestingly, the hotspots manage to indicate which part of the spine is affected by scoliosis; in Fig. 8, we can see hotspots examples of thoracic scoliosis which localized around the thoracic region (upper spine) and examples of lumbar scoliosis which localized around the lumbar region (lower spine).
Severity of Scoliosis. The output prediction of the network, specifically scoliosis, can be interpreted as a soft score of the task (softmax of the last layer). Since the ease of predicting scoliosis directly relates to the how curved the spine is, the more confident the network is about the prediction, the more likely that the scan has scoliosis. Figure 9 shows scans on the test set alongside their soft scores. This soft score of scoliosis can be used to monitor disease progression of patients with scoliosis, where getting higher scores across a period of time i.e. a longitudinal study of the subject would mean the scoliosis is getting worse.
6 Conclusion
We have shown that scoliosis can be predicted automatically via DXA scans, and that predictions can improved by adding more supervision in the form of intermediate representations, which in our case comes in the form of a soft segmentation mask of the spine and other body parts. We have also demonstrated that the evidence for the scoliosis classification can be weakly localized as hot spots, and that the score defines a grading for scoliosis severity. One possible future work is to predict the direction of the apex of the curves.
References
Asher, M.A., Burton, D.C.: Adolescent idiopathic scoliosis: natural history and long term treatment effects. Scoliosis 1(1), 2 (2006)
Burkhart, T.A., Arthurs, K.L., Andrews, D.M.: Manual segmentation of DXA scan images results in reliable upper and lower extremity soft and rigid tissue mass estimates. J. Biomech. 42(8), 1138–1142 (2009)
Chan, T.F., Vese, L.A.: Active contours without edges. IEEE Trans. Image Process 10(2), 266–277 (2001). https://doi.org/10.1109/83.902291
Chatfield, K., Simonyan, K., Vedaldi, A., Zisserman, A.: Return of the devil in the details: delving deep into convolutional nets. In: Proceedings of BMVC (2014)
Clark, E.M., Tobias, J.H., Fairbank, J.: The impact of small spinal curves in adolescents who have not presented to secondary care: a population-based cohort study. Spine 41(10), E611–617 (2016)
Jamaludin, A., Kadir, T., Zisserman, A.: SpineNet: automatically pinpointing classification evidence in spinal MRIs. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 166–175. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_20
Jamaludin, A., Kadir, T., Zisserman, A.: SpineNet: automated classification and evidence visualization in spinal MRIs. Med. Image Anal. 41, 63–73 (2017)
Pehrsson, K., Bake, B., Larsson, S., Nachemson, A.: Lung function in adult idiopathic scoliosis: a 20 year follow up. Thorax 46(7), 474–478 (1991)
Roberts, M.G., Pacheco, E.M., Mohankumar, R., Cootes, T.F., Adams, J.E.: Detection of vertebral fractures in DXA VFA images using statistical models of appearance and a semi-automatic segmentation. Osteoporos. Int. 21(12), 2037–2046 (2010)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Shepherd, J.A., Ng, B.K., Fan, B., Schwartz, A.V., Cawthon, P., Cummings, S.R., Kritchevsky, S., Nevitt, M., Santanasto, A., Cootes, T.F.: Modeling the shape and composition of the human body using dual energy X-ray absorptiometry images. PLoS ONE 12(4), e0175857 (2017)
Taylor, H.J., et al.: Identifying scoliosis in population-based cohorts: development and validation of a novel method based on total-body dual-energy x-ray absorptiometric scans. Calcif. Tissue Int. 92(6), 539–547 (2013)
Vedaldi, A., Lenc, K.: MatConvNet: convolutional neural networks for MATLAB. In: Proceedings of ACMM (2015)
Wu, J., Wang, Y., Xue, T., Sun, X., Freeman, W.T., Tenenbaum, J.B.: MarrNet: 3D shape reconstruction via 2.5D sketches. In: Advances in Neural Information Processing Systems (2017)
Wu, J., et al.: Single image 3D interpreter network. In: Leibe, B., Matas, J., Sebe, N., Welling, M. (eds.) ECCV 2016. LNCS, vol. 9910, pp. 365–382. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46466-4_22
Zhang, J., Bargal, S.A., Lin, Z., Brandt, J., Shen, X., Sclaroff, S.: Top-down neural attention by excitation backprop. Int. J. Comput. Vis. 126, 1084–1102 (2017). https://doi.org/10.1007/s11263-017-1059-x
Acknowledgements
We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. The UK Medical Research Council and the Wellcome Trust (Grant ref: 102215/2/13/2) and the University of Bristol provide core support for ALSPAC. This publication is the work of the authors and Amir Jamaludin will serve as guarantor for the contents of this paper. This research was specifically funded by the British Scoliosis Research Foundation, and the DXA scans were funded through the Wellcome Trust (grants 084632 and 079960).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Jamaludin, A., Kadir, T., Clark, E., Zisserman, A. (2019). Predicting Scoliosis in DXA Scans Using Intermediate Representations. In: Zheng, G., Belavy, D., Cai, Y., Li, S. (eds) Computational Methods and Clinical Applications for Spine Imaging. CSI 2018. Lecture Notes in Computer Science(), vol 11397. Springer, Cham. https://doi.org/10.1007/978-3-030-13736-6_2
Download citation
DOI: https://doi.org/10.1007/978-3-030-13736-6_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-13735-9
Online ISBN: 978-3-030-13736-6
eBook Packages: Computer ScienceComputer Science (R0)