Abstract
Coronavirus genomes are single-stranded positive-sense RNA that are transcribed into a nested set of 3′ coterminal subgenomic RNAs for gene expression. Members of the Coronaviridae express canonical polymerase genes, as well as structural genes, including S, E, M, and N, but also express a highly divergent set of accessory genes whose open reading frames are interspersed among the structural genes within the 3′ one-third of the viral genome. The accessory genes are thought to contain “luxury” functions that are often not required for in-vitro virus replication. The severe acute respiratory syndrome coronavirus (SARS-CoV) expresses eight such accessory genes (ORF3a, -3b, -6, -7a, -7b, -8a, -8b, and -9b), the most of any known coronavirus. This chapter will review our current knowledge of expression, structure, and function of each of the SARS-CoV accessory genes.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
- Accessory Gene
- Severe Acute Respiratory Syndrome
- Syrian Golden Hamster
- Mouse Hepatitis Virus
- Severe Acute Respiratory Syndrome Coronavirus
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
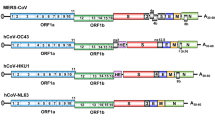
Coronaviruses have large, positive-stranded, RNA genomes ranging from 27 to 31 kb in size (Fig. 10.1). The 3′ end of the genome contains open reading frames (ORFs) encoding for canonical structural proteins including envelope (E), membrane (M), spike (S), and the nucleocapsid protein (N). Interspersed among the structural genes are novel ORFs that encode additional virus-specific proteins. The novel ORFs have been aptly renamed “accessory genes.” Coronavirus genomes differ greatly in number, location, and size of the accessory genes; as detailed in Fig. 10.1, viruses such as hCoV-NL63 can express as few as one, or in the case of SARS-CoV and the recently identified whale coronavirus (Mihindukulasuriya et al. 2008), as many as eight accessory proteins.
Genomic diversity among coronaviruses. Genomic organization of representative virus strains from groups I, II, and III coronaviruses are detailed. Relative locations of each replicase, structural, and accessory ORF are highlighted for each virus, with primary ORFs on respective subgenomic RNAs in black boxes and downstream ORFs in multicistronic genes shaded in gray. The Genbank accession ID and genome size for each annotated strain is displayed and nucleotide length of each ORF is shown
Three independent reports simultaneously detailed the SARS-CoV genomic sequence and annotations (Marra et al. 2003; Rota et al. 2003; Thiel et al. 2003), which led to some confusion regarding the nomenclature of the accessory genes. Annotations by Thiel et al. were the least stringent and identified the eight currently accepted accessory genes; additionally, nomenclature consistent with other coronaviruses was given to them which will be used throughout this chapter. Numerous studies have reported on the expression of all eight SARS-CoV accessory genes. Few polymorphisms have been identified in the more than 100 sequenced SARS-CoV isolates, suggesting a selective pressure to maintain their expression. This chapter will review current data regarding the expression, structure, function, and molecular biology of the SARS-CoV accessory genes (Table 10.1).
2 SARS-CoV Accessory Gene Expression and Function
2.1 ORF3a and ORF3b
ORF3a is the largest of the SARS-CoV accessory proteins at 274 amino acids in length with a native molecular weight of 31 kDa (Tan et al. 2004c). ORF3a is expressed from the 5′-most ORF present in sgRNA3, which contains a minimal transcriptional regulatory sequence (TRS, 5′-ACGAAC-3′) (Snijder et al. 2003) immediately upstream of the translation initiation codon. Expression has been confirmed in virus-infected cells (Tan et al. 2004c; Yu et al. 2004; Zeng et al. 2004) and antibodies recognizing ORF3a have been detected in SARS-CoV patient convalescent sera (Guo et al. 2004; Tan et al. 2004b; Yu et al. 2004; Zeng et al. 2004; Qiu et al. 2005; Yount et al. 2005; Zhong et al. 2005).
ORF3a is a hydrophobic, triple membrane-spanning protein with an N-terminal ectodomain and a C-terminal intracellular domain (Tan et al. 2004c; Lu et al. 2006) and is predicted to contain an amino-terminal signal sequence spanning residues 1–16 (Law et al. 2005). ORF3a is O-glycosylated (Oostra et al. 2006) and localizes predominantly to the Golgi and plasma membrane (Tan et al. 2004c; Yu et al. 2004; Ito et al. 2005; Yuan et al. 2005a; Lu et al. 2006; von Brunn et al. 2007). Plasma membrane trafficking and endocytosis is mediated by a YxxΦ motif and a diacidic motif in the cytoplasmic tail that are juxtaposed to one another within a 14 amino acid region (Tan et al. 2004c). This region of the cytoplasmic tail has also been shown to mediate G1 phase cell cycle arrest in HEK-293T cells transfected with ORF3a cDNA (Yuan et al. 2007). No evidence has been presented regarding control of cell cycle progression in SARS-CoV infected cells, so the functional significance of this observation is unknown.
Analysis of virus-like particles and purified virions demonstrated that ORF3a is packaged into virions (Zeng et al. 2004; Ito et al. 2005; Shen et al. 2005). Potential interactions between ORF3a and the viral proteins S, E, M, and ORF7a have been demonstrated (Tan et al. 2004c; Zeng et al. 2004). Antibodies specific to ORF3a have shown the surprising ability to neutralize virus infection in multiple ways. Because some ORF3a is trafficked to the cell surface, antibodies recognizing the amino-terminal extracellular domain initiate complement-mediated lysis in the presence of functional complement proteins (Zhong et al. 2006). ORF3a N-terminus-specific antibodies are also capable of neutralizing infectious virus in microneutralization assays (Akerstrom et al. 2006). Although the precise mechanism of virus neutralization is unknown, these studies suggest that ORF3a may be a potential vaccine target.
ORF3a forms homodimers and homotetramers via disulfide linkages and the oligomeric complexes have been proposed to function as potassium-permeable ion channels in Xenopus oocytes (Lu et al. 2006). The authors propose a role for ORF3a in virus release; however, an ORF3a deletion virus is not defective in virus entry, replication, assembly, or budding (Yount et al. 2005). Additional evidence for ORF3a involvement in the virus lifecycle stems from the observation that ORF3a directly binds the 5′ untranslated region (UTR) of the viral genome, mediated by residues 125–200 of the cytoplasmic tail (Sharma et al. 2007). ORF3a is not required for virus replication in vitro or in a small animal model, as a recombinant SARS-CoV strain deleted of ORF3a replicates to wild-type levels in numerous cell lines and in BALB/c mice (Yount et al. 2005). Like many other SARS-CoV viral proteins, transient expression of ORF3a has been shown to induce apoptosis in Vero cells (Law et al. 2005). Expression of ORF3a in 293T cells can also augment NF-κB activity (Kanzawa et al. 2006; Narayanan et al. 2007) and expression in A549 cells upregulates the expression and secretion of fibrinogen (Tan et al. 2005).
ORF3b is also expressed from sgRNA3 with a coding sequence overlapping both the ORF3a and E sequences. Although present in the genomes of human and civet SARS-CoV isolates, the ORF3b open reading frame is not present in bat SARS-CoV isolates due to a stop codon early in the ORF3b sequence (Ren et al. 2006). The ORF3b protein is 154 amino acids in length and the translation initiation AUG codon lies 418 nucleotides downstream of the ORF3a AUG. No transcriptional regulatory sequence exists upstream of the ORF3b sequence and there are 11 AUG sequences between the ORF3a and ORF3b initiation codons which likely precludes leaky scanning as the mechanism of ORF3b translation. Although the ORF3b coding sequence lies in a translationally unfavorable context, the protein has been observed in virus-infected Vero cells (Chan et al. 2005) and antibodies recognizing ORF3b have also been found in patient convalescent sera (Guo et al. 2004; Chow et al. 2006).
Bioinformatics analysis predicted two nuclear localization signals (NLS) within the ORF3b protein and subsequent analysis has confirmed the nuclear and nucleolar localization of ORF3b (Pewe et al. 2005; Yuan et al. 2005c; Kopecky-Bromberg et al. 2007; von Brunn et al. 2007). A separate report has also suggested ORF3b localizes to mitochondria (Yuan et al. 2006a). No signal peptides or transmembrane domains (TMD) are predicted, suggesting ORF3b is an 18 kDa soluble protein.
ORF3b induces both apoptosis and necrosis in multiple cell lines (Yuan et al. 2005b; Khan et al. 2006) and prevents cell cycle transition from G0/G1 to S phase (Yuan et al. 2005b). Again, cell cycle regulation has not been identified in SARS-CoV infected cells, so the biological relevance of this finding is unknown. ORF3b has been shown to be an interferon antagonist (Kopecky-Bromberg et al. 2007). SARS-CoV has the ability to prevent interferon-β (IFN-β) production in infected cells (Spiegel et al. 2005) and ORF3b may be one of several viral proteins whose function is to prevent IFN-β induction. Like ORF6, the ORF3b protein prevents IFN-β expression by inhibiting the function of interferon regulatory factor 3 (IRF-3) and also prevents transcription from interferon stimulated response element (ISRE)-containing promoters.
2.2 ORF6
ORF6 is a 7.5 kDa protein of 63 amino acids and the only ORF translated from sgRNA6. ORF6 expression has been confirmed in lung and enteric tissue from infected patients (Chan et al. 2005; Geng et al. 2005). No signal peptide is predicted within ORF6 and the N-terminal 40 amino acids are predominantly hydrophobic, with the exception of six charged residues spaced approximately seven amino acids apart, suggesting an amphipathic alpha-helical structure (Netland et al. 2007). The C-terminal 23 amino acids are largely composed of hydrophilic, charged residues. The hydrophobic N-terminus of ORF6 mediates membrane association (Pewe et al. 2005); however, the evenly spaced charged residues suggest that the hydrophobic region may not form a genuine transmembrane domain. In agreement with this, Netland et al. identified that both the N- and C-termini are cytoplasmic (Netland et al. 2007).
ORF6 is packaged into virus particles and incorporated into virus-like particles; the protein was also released from HEK-293T cells into the supernatant when expressed independently of other viral proteins (Huang et al. 2007). The packaging of ORF6 into virions is likely due to specific interactions with other structural protein(s), as ORF6 is not present in purified mouse hepatitis virus (MHV) particles collected from cells coexpressing SARS-CoV ORF6 (Pewe et al. 2005; Tangudu et al. 2007). Direct interaction between ORF6 and the nonstructural protein nsp8 has been observed, suggesting that ORF6 may play a role in virus assembly or replication processes (Kumar et al. 2007). Given the subcellular localization of ORF6 to the approximate area of viral RNA replication and virus assembly, further studies analyzing contributions of ORF6 to virus replication are warranted.
Heterologous expression of ORF6 by an attenuated MHV variant resulted in increased virulence and higher viral titers in C57BL/6 mice (Pewe et al. 2005). Subsequently, Kopecky-Bromberg et al. (2007) identified that ORF6 antagonizes IFN-β production from cells infected with Sendai virus by inhibiting IRF-3 activation. ORF6 was also able to inhibit the nuclear translocation of STAT1 within IFN-β treated cells. Frieman et al. (2007) demonstrated that ORF6 interacts with the nuclear import factor karyopherin-α2 (KPNA2) at ER/Golgi membranes, sequestering KPNA2 and karyopherin-β1 complexes resulting in loss of STAT1 translocation into the nucleus and subsequent inhibition of STAT1-activated antiviral genes.
Deletion of ORF6 in a recombinant SARS-CoV was initially described as having little to no effect on virus replication in vitro or in the BALB/c mouse model of virus replication (Yount et al. 2005). In contrast, the deletion of ORF6 did result in subtle changes to SARS-CoV replication after low multiplicity of infection (MOI) infection of Vero cells (Zhao et al. 2009) and in virus virulence and replication in the human ACE2-transgenic C57BL/6 mouse model (Zhao et al. 2009).
2.3 ORF7a and ORF7b
Gene 7 contains two ORFs, ORF7a and ORF7b. The ORF7a translation initiation sequence lies immediately juxtaposed to the 5′ TRS and translation results in a 122 amino acid protein approximately 17.5 kDa in size prior to cleavage of an N-terminal signal sequence (Fielding et al. 2004; Nelson et al. 2005). ORF7a is highly conserved among all sequenced human and animal SARS-CoV isolates including bat SARS-CoV and its expression has been detected in lung and enteric tissues from infected patients (Chan et al. 2005; Chen et al. 2005).
ORF7a is a type-I integral membrane protein containing a 15 amino acid N-terminal signal sequence, 81 residue luminal domain, 21 residue TMD, and a 5 amino acid cytoplasmic tail. The crystal structure of the luminal domain has been solved, revealing a seven-stranded beta sandwich that adopts a compact Ig-like fold (Nelson et al. 2005). The amino acid sequence contains little homology to any other known protein but the luminal domain structure does have some similarity to the D1 domain of human ICAM-1 (Nelson et al. 2005; Hanel and Willbold 2007).
ORF7a localizes predominantly to the Golgi region of transfected and infected cells (Nelson et al. 2005; Kopecky-Bromberg et al. 2006; Pekosz et al. 2006; Schaecher et al. 2007a,b). The short cytoplasmic tail contains a dibasic ER export motif (RK) × (RK) that mediates export from the ER through interactions with COPII machinery (Nelson et al. (2005) and our unpublished data) and the Golgi retention motif resides within the TMD and cytoplasmic tail (Nelson et al. 2005). ORF7a has also been observed in purified virus particles and virus-like particles (VLPs) (Huang et al. 2006).
The precise function of ORF7a in virus infected cells remains unclear. Numerous studies have reported proapoptotic effects of ORF7a when expressed via cDNA transfection (Tan et al. 2004a, 2007; Kopecky-Bromberg et al. 2006; Yuan et al. 2006b; Schaecher et al. 2007b). In-vitro expression of ORF7a results in apoptosis through a caspase-dependent pathway, inhibition of cellular protein synthesis, blockage of cell cycle progression at G0/G1 phase, activation of NF-κB, increased IL-8 promoter activity, and activation of p38 MAP kinase (Tan et al. 2004a, 2007; Kanzawa et al. 2006; Kopecky-Bromberg et al. 2006; Yuan et al. 2006b; Schaecher et al. 2007b). These varied results all suggest a role for ORF7a in altering the host cellular environment. A recombinant SARS-CoV lacking gene 7 induces early-stage apoptosis and cell death similar to wild-type virus, but latter stages of the apoptotic cascade are altered as oligonucleosomal DNA fragmentation is significantly reduced compared to cells infected with wild-type virus (Schaecher et al. 2007b); however, gene 7 deletion viruses have no discernable defect in virus replication, pathogenesis, or lethality in wild-type or immunodeficient Syrian golden hamsters (Schaecher et al. 2007b; Schaecher et al. 2008b) or in Balb/c mice (Yount et al. 2005).
Several studies have identified potential interactions between cellular proteins and ORF7a. Tan et al. have reported that the ORF7a transmembrane domain mediates interaction with the antiapoptotic cellular protein BCL-XL (Tan et al. 2007). Although ORF7a does not localize to mitochondria, it is possible that direct interaction with BCL-XL occurs at the ER membranes. This could tilt the balance of pro- and antiapoptotic regulators at the mitochondria resulting in activation of a cell death cascade. Our preliminary data has not revealed any detectable colocalization between ORF7a and BCL-XL at any subcellular site (data not shown), so further studies are necessary to analyze the potential interactions of ORF7a with BCL-XL.
Potential interactions between ORF7a and the small glutamine-rich tetraicopeptide repeat-containing protein hSGT were identified using a two-hybrid screen (Fielding et al. 2006); residues 1–96 of ORF7a were determined to be the interacting region. These residues include the signal peptide (which is not present in the mature protein), and the ORF7a ectodomain domain which is in the Golgi lumen (Nelson et al. 2005). Since hSGT is a cytosolic protein, it is unclear how this interaction can occur in mammalian cells.
Interactions between ORF7a and a third cellular protein, lymphocyte function-associated antigen 1 (LFA-1), have been identified (Hanel and Willbold 2007). LFA-1 is expressed exclusively on leukocytes and is involved in lymphocyte homing and intercellular interactions during inflammatory immune responses. Recombinant ORF7a binds to the extracellular domain of LFA-1; however, the significance of this is unknown as ORF7a is not secreted from expressing cells, nor is it present on the plasma membrane of infected cells (Nelson et al. 2005; Huang et al. 2006).
The ORF7b protein is encoded by an ORF beginning 365 nucleotides from the sgRNA7 TRS. The start codon for ORF7b overlaps but is out of frame with the stop codon for ORF7a and translation occurs via ribosome leaky scanning (Schaecher et al. 2007a). ORF7b localizes to the Golgi region of cDNA transfected and SARS-CoV infected cells (Pekosz et al. 2006; Schaecher et al. 2007a,b). The ORF7b protein is a highly hydrophobic protein of 44 amino acids and a molecular weight of 5.5 kDa. ORF7b has no predicted signal sequence and is an integral membrane protein containing a 22 amino acid transmembrane domain and a cytoplasmic C-terminus (Schaecher et al. 2007b), suggesting it is a type-III integral membrane protein. The Golgi localization motif is present within the ORF7b TMD and can confer Golgi localization to a protein that is normally found on the plasma membrane (Schaecher et al. 2008a).
The ORF7b protein is associated with intracellular virus particles and is present in purified virions (Schaecher et al. 2007a). Expression of ORF7b induces apoptosis in transfected cells, but to a lesser extent than ORF7a (Kopecky-Bromberg et al. 2006; Schaecher et al. 2007b). Like the other SARS-CoV accessory proteins, however, it is not required for replication in vitro, in mice, or in Syrian golden hamsters (Sims et al. 2005; Yount et al. 2005; Schaecher et al. 2007b; Schaecher et al. 2008b).
2.4 ORF8a and ORF8b
SARS-CoV strains isolated from civet cats, raccoon dogs, and bats in China as well as early human isolates contain a single, continuous ORF8 sequence referred to as ORF8ab. Virus isolated from middle and late stages of the human outbreak, comprising the majority of human isolates, was found to have a 29-nucleotide deletion within the ORF creating two overlapping reading frames designated ORF8a and ORF8b (Fig. 10.1) (Guan et al. 2003; Chinese 2004; Lau et al. 2005). The ORF8a/8ab translation initiation codon lies proximal to the strong sgRNA8 TRS, and translation of full length ORF8ab results in a 122 amino acid protein with an N-terminus identical to that of ORF8a, and C-terminus identical to that of ORF8b. Translation of ORF8a and ORF8b, however, results in products of 39 and 84 residues, respectively. It has been suggested that the ORF8ab protein is a functional protein that was lost upon transmission to humans, and the resultant ORF8a and/or ORF8b proteins are likely nonfunctional (Snijder et al. 2003; Oostra et al. 2007).
The N-terminus of both the large ORF8ab protein and truncated ORF8a contains a predicted hydrophobic signal sequence. ORF8ab colocalizes with ER markers and is membrane-associated; it is presumed that ORF8ab is not an integral membrane protein and exists either as a soluble protein in the lumen ER lumen or is peripherally associated with the luminal face of ER membranes (Oostra et al. 2007).
The ORF8a translation product is a small protein of 5.3 kDa and antibodies recognizing ORF8a have been detected in patient convalescent sera (Chen et al. 2007). An ORF8a–GFP fusion protein colocalizes with the ER marker calnexin (Oostra et al. 2007); however, the 39 amino acid ORF8a is too small to cotranslationally interact with the signal recognition particle (SRP) and is likely released into the cytosol prior to translocation. The cytosolic accumulation of ORF8a in SARS-CoV infected Vero cells supports this theory (Keng et al. 2006), although ORF8a localization to mitochondria has also been described where it may impart a proapoptotic effect (Chen et al. 2007). Little is known regarding ORF8a function in the virus lifecycle; however, a dose-dependent increase in virus replication has been observed in stable cell lines expressing varying levels of ORF8a (Keng et al. 2006).
No TRS exists upstream of ORF8b and translation initiation most likely does not involve a conventional ribosomal scanning mechanism, as scanning ribosomes would have to pass two AUG sequences prior to reaching the predicted ORF8b initiation codon. Contradictory evidence regarding the expression of ORF8b exists. Keng et al. (2006) have demonstrated that ORF8b is expressed in SARS-CoV infected Vero cells. However, two independent reports have suggested that ORF8b is not expressed from the ORF8a/b message either in SARS-CoV infected cells or ORF8a/b cDNA-transfected cells (Le et al. 2007; Oostra et al. 2007). When expressed independently, ORF8b localization is distributed evenly throughout the cytoplasm (Keng et al. 2006; Le et al. 2007).
Little is known regarding the function of ORF8b in virus-infected cells. Cotransfection of ORF8b and SARS-CoV E protein results in a posttranslational downregulation of E, a phenomenon not observed in cells coexpressing E and ORF8ab (Keng et al. 2006). Direct interactions between ORF8b and E were detected suggesting a potential role for ORF8b in modulating degradation or stability of E. Further analysis of ORF8b expression and function is required to confirm whether ORF8b is in fact translated and functional in SARS-CoV infected cells.
2.5 ORF9b
Some group 2 coronaviruses encode a protein in an alternative reading frame contained entirely within the N gene, termed I or “internal” (Fig. 10.1). SARS-CoV also encodes an internal ORF, termed ORF9b. The predicted initiation codon lies ten nucleotides from the nucleocapsid initiation AUG codon with no intervening AUG sequences, providing a prime opportunity for ribosomal leaky scanning mediated translation initiation. Antibodies specific to ORF9b have been detected in patient sera (Guo et al. 2004; Qiu et al. 2005; Zhong et al. 2005) and protein was detected in tissues from SARS-CoV infected patients (Chan et al. 2005), suggesting that ORF9b is expressed during virus infection.
The ORF9b protein is 98 amino acids in length and has a molecular weight of approximately 11 kDa. The crystal structure of the protein reveals an intertwined symmetrical dimer whose β-sheets form a tent-like structure (Meier et al. 2006). Mass spectroscopy analysis revealed a long fatty acid or fatty acid ester molecule present in the structure’s hydrophobic tunnel; it has been speculated that the ORF9b protein may anchor to intracellular membranes by internalizing one or more lipidic tails from membrane lipids (Meier et al. 2006). ORF9b localizes to the ER region of transfected cells, suggesting that the protein may interact with intracellular membranes although no transmembrane regions are predicted within ORF9b. Yeast-two-hybrid experiments have suggested that ORF9b interacts with viral proteins nsp8, nsp14, and ORF7b, and provide further evidence of homotypic interactions (von Brunn et al. 2007); however, no functional analysis has been described to date regarding the role of ORF9b in the context of the virus replication cycle.
3 Conclusions
The large genetic diversity among coronaviruses raises questions regarding the origins of the accessory genes. Have they been acquired through horizontal transfer from host cellular genetic material or heterologous viruses? Have they been acquired through gene duplication and subsequent mutation within the virus’s own genome? Evidence has been presented pointing to both possibilities. For example, the group 2 coronavirus hemagglutinin esterase (HE) genes have approximately 30% amino acid identity with the influenza C virus (ICV) HE-1 subunit of the HEF gene; it has been suggested that an early group 2 coronavirus captured HE from ICV during a mixed infection (Luytjes et al. 1988). For the latter theory, several of the SARS-CoV accessory genes may have originated from internal duplication, fusion, or shifting events (Inberg and Linial 2004). ORF3a may have arisen through gene duplication and mutation of the M ORF (Inberg and Linial 2004; Masters 2006; Oostra et al. 2006). Furthermore, ORF7b and ORF8a have some amino acid sequence similarity to ORF3a, suggesting that the larger ORF3a may have given rise to ORF7b and ORF8 (Inberg and Linial 2004). Regardless of the origins of the SARS-CoV accessory genes, the severe disease caused by the virus highlights the need to study the functions of its eight accessory genes.
The observation that the SARS-CoV accessory proteins are dispensable for virus replication in vitro is not uncommon among coronaviruses (Yount et al. 2005). It is striking that a virus lacking each of the accessory proteins, either individually or in combination, replicates efficiently in BALB/c mice. This finding may indicate that other animal models for the in-vivo analysis of accessory gene function are required. It is also possible that some or all of the accessory genes play a more important role during infection of humans, bats or other animal reservoirs of the virus. Identifying the functions of these accessory proteins may prove beneficial not only for further understanding of SARS-CoV pathogenesis, but also for better understanding and preparedness for future viral infectious diseases.
References
Akerstrom S, Tan YJ et al (2006) Amino acids 15–28 in the ectodomain of SARS coronavirus 3a protein induces neutralizing antibodies. FEBS Lett 580(16):3799–3803
Chan WS, Wu C et al (2005) Coronaviral hypothetical and structural proteins were found in the intestinal surface enterocytes and pneumocytes of severe acute respiratory syndrome (SARS). Mod Pathol 18:1432–1439
Chen YY, Shuang B et al (2005) The protein X4 of severe acute respiratory syndrome-associated coronavirus is expressed on both virus-infected cells and lung tissue of severe acute respiratory syndrome patients and inhibits growth of Balb/c 3 T3 cell line. Chin Med J 118(4):267–274
Chen CY, Ping YH et al (2007) Open reading frame 8a of the human severe acute respiratory syndrome coronavirus not only promotes viral replication but also induces apoptosis. J Infect Dis 196(3):405–415
Chinese SMEC (2004) Molecular evolution of the SARS Coronavirus during the course of the SARS epidemic in China. Science 303(5664):1666–1669
Chow SC, Ho CY et al (2006) Specific epitopes of the structural and hypothetical proteins elicit variable humoral responses in SARS patients. J Clin Pathol 59(5):468–476
Fielding BC, Tan Y-J et al (2004) Characterization of a unique group-specific protein (U122) of the severe acute respiratory syndrome Coronavirus. J Virol 78(14):7311–7318
Fielding BC, Gunalan V et al (2006) Severe acute respiratory syndrome coronavirus protein 7a interacts with hSGT. Biochem Biophys Res Commun 343(4):1201–1208
Frieman M, Yount B et al (2007) Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J Virol 81(18):9812–9824
Geng H, Liu Y-M et al (2005) The putative protein 6 of the severe acute respiratory syndrome-associated coronavirus: expression and functional characterization. FEBS Lett 579(30):6763–6768
Guan Y, Zheng BJ et al (2003) Isolation and characterization of viruses related to the SARS Coronavirus from animals in Southern China. Science 302(5643):276–278
Guo JP, Petric M et al (2004) SARS corona virus peptides recognized by antibodies in the sera of convalescent cases. Virology 324(2):251–256
Hanel K, Willbold D (2007) SARS-CoV accessory protein 7a directly interacts with human LFA-1. Biol Chem 388(12):1325–1332
Huang C, Ito N et al (2006) Severe acute respiratory syndrome coronavirus 7a accessory protein is a viral structural protein. J Virol 80(15):7287–7294
Huang C, Peters CJ et al (2007) Severe acute respiratory syndrome coronavirus accessory protein 6 is a virion-associated protein and is released from 6 protein-expressing cells. J Virol 81(10):5423–5426
Inberg A, Linial M (2004) Evolutional insights on uncharacterized SARS coronavirus genes. FEBS Lett 577(1–2):159–164
Ito N, Mossel EC et al (2005) Severe acute respiratory syndrome Coronavirus 3a protein is a viral structural protein. J Virol 79(5):3182–3186
Kanzawa N, Nishigaki K et al (2006) Augmentation of chemokine production by severe acute respiratory syndrome coronavirus 3a/X1 and 7a/X4 proteins through NF-[kappa]B activation. FEBS Lett 580(30):6807–6812
Keng C-T, Choi Y-W et al (2006) The human severe acute respiratory syndrome coronavirus (SARS-CoV) 8b protein is distinct from its counterpart in animal SARS-CoV and down-regulates the expression of the envelope protein in infected cells. Virology 354(1):132–142
Khan S, Fielding BC et al (2006) Over-expression of severe acute respiratory syndrome coronavirus 3b protein induces both apoptosis and necrosis in Vero E6 cells. Virus Res 122(1–2):20–27
Kopecky-Bromberg SA, Martinez-Sobrido L et al (2006) 7a protein of severe acute respiratory syndrome Coronavirus inhibits cellular protein synthesis and activates p38 mitogen-activated protein kinase. J Virol 80(2):785–793
Kopecky-Bromberg SA, Martinez-Sobrido L et al (2007) Severe acute respiratory syndrome Coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol 81(2):548–557
Kumar P, Gunalan V et al (2007) The nonstructural protein 8 (nsp8) of the SARS coronavirus interacts with its ORF6 accessory protein. Virology 366(2):293–303
Lau SKP, Woo PCY et al (2005) Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. PNAS 102(39):14040–14045
Law PTW, Wong C-H et al (2005) The 3a protein of severe acute respiratory syndrome-associated coronavirus induces apoptosis in Vero E6 cells. J Gen Virol 86(7):1921–1930
Le TM, Wong HH et al (2007) Expression, post-translational modification and biochemical characterization of proteins encoded by subgenomic mRNA8 of the severe acute respiratory syndrome coronavirus. FEBS J 274(16):4211–4222
Lu W, Zheng B-J et al (2006) Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. PNAS 103(33):12540–12545
Luytjes W, Bredenbeek PJ et al (1988) Sequence of mouse hepatitis virus A59 mRNA 2: indications for RNA recombination between coronaviruses and influenza C virus. Virology 166(2):415–422
Marra MA, Jones SJM et al (2003) The genome sequence of the SARS-associated Coronavirus. Science 300(5624):1399–1404
Masters PS (2006) The molecular biology of coronaviruses. Adv Virus Res 66:193–292
Meier C, Aricescu AR et al (2006) The crystal structure of ORF-9b, a lipid binding protein from the SARS Coronavirus. Structure 14(7):1157–1165
Mihindukulasuriya KA, Wu G, et al (2008) Identification of a novel Coronavirus from a Beluga Whale using a pan-viral microarray. J Virol 82(10):5084–5088
Narayanan K, Huang C, et al (2007) SARS coronavirus accessory proteins. Virus Res 133(1):113–121
Nelson CA, Pekosz A et al (2005) Structure and intracellular targeting of the SARS-Coronavirus Orf7a accessory protein. Structure 13(1):75–85
Netland J, Ferraro D et al (2007) Enhancement of murine coronavirus replication by severe acute respiratory syndrome coronavirus protein 6 requires the N-terminal hydrophobic region but not C-terminal sorting motifs. J Virol 81(20):11520–11525
Oostra M, de Haan CAM et al (2006) Glycosylation of the severe acute respiratory syndrome coronavirus triple-spanning membrane proteins 3a and M. J Virol 80(5):2326–2336
Oostra M, de Haan CA et al (2007) The 29-nucleotide deletion present in human but not in animal severe acute respiratory syndrome coronaviruses disrupts the functional expression of open reading frame 8. J Virol 81(24):13876–13888
Pekosz A, Schaecher SR et al (2006) Structure, expression, and intracellular localization of the SARS-CoV accessory proteins 7a and 7b. Adv Exp Med Biol 581:115–120
Pewe L, Zhou H et al (2005) A severe acute respiratory syndrome-associated coronavirus-specific protein enhances virulence of an attenuated murine coronavirus. J Virol 79(17):11335–11342
Qiu M, Shi Y et al (2005) Antibody responses to individual proteins of SARS coronavirus and their neutralization activities. Microbes Infect 7(5–6):882–889
Ren W, Li W et al (2006) Full-length genome sequences of two SARS-like coronaviruses in horseshoe bats and genetic variation analysis. J Gen Virol 87(11):3355–3359
Rota PA, Oberste MS et al (2003) Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300(5624):1394–1399
Schaecher SR, Mackenzie JM et al (2007a) The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. J Virol 81(2):718–731
Schaecher SR, Touchette E et al (2007b) Severe acute respiratory syndrome coronavirus gene 7 products contribute to virus-induced apoptosis. J Virol 81(20):11054–11068
Schaecher SR, Diamond MS et al (2008a) The transmembrane domain of the severe acute respiratory syndrome coronavirus ORF7b protein is necessary and sufficient for its retention in the Golgi complex. J Virol 82(19):9477–9491
Schaecher SR, Stabenow J et al (2008b) An immunosuppressed Syrian golden hamster model for SARS-CoV infection. Virology 380(2):312–321
Sharma K, Surjit M et al (2007) The 3a accessory protein of SARS coronavirus specifically interacts with the 5'UTR of its genomic RNA, using a unique 75 amino acid interaction domain. Biochemistry 46(22):6488–6499
Shen S, Lin PS et al (2005) The severe acute respiratory syndrome coronavirus 3a is a novel structural protein. Biochem Biophys Res Commun 330(1):286–292
Sims AC, Baric RS et al (2005) Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J Virol 79(24):15511–15524
Snijder EJ, Bredenbeek PJ et al (2003) Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol 331(5):991–1004
Spiegel M, Pichlmair A et al (2005) Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J Virol 79(4):2079–2086
Tan Y-J, Fielding BC et al (2004a) Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J Virol 78(24):14043–14047
Tan Y-J, Goh P-Y et al (2004b) Profiles of antibody responses against severe acute respiratory syndrome coronavirus recombinant proteins and their potential use as diagnostic markers. Clin Diagn Lab Immunol 11(2):362–371
Tan Y-J, Teng E et al (2004c) A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J Virol 78(13):6723–6734
Tan Y-J, Tham P-Y et al (2005) The severe acute respiratory syndrome coronavirus 3a protein up-regulates expression of fibrinogen in lung epithelial cells. J Virol 79(15):10083–10087
Tan YX, Tan TH et al (2007) The induction of apoptosis by the severe acute respiratory syndrome (SARS)-Coronavirus 7a protein is dependent on its interaction with the Bcl-XL protein. J Virol 81(12):6346–6355
Tangudu C, Olivares H et al (2007) Severe acute respiratory syndrome coronavirus protein 6 accelerates murine coronavirus infections. J Virol 81(3):1220–1229
Thiel V, Ivanov KA et al (2003) Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol 84(9):2305–2315
von Brunn A, Teepe C et al (2007) Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS ONE 2(5):e459
Yount B, Roberts RS et al (2005) Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J Virol 79(23):14909–14922
Yu CJ, Chen YC et al (2004) Identification of a novel protein 3a from severe acute respiratory syndrome coronavirus. FEBS Lett 565(1–3):111–116
Yuan X, Li J et al (2005a) Subcellular localization and membrane association of SARS-CoV 3a protein. Virus Res 109(2):191–202
Yuan X, Shan Y et al (2005b) G0/G1 arrest and apoptosis induced by SARS-CoV 3b protein in transfected cells. Virol J 2(66):66
Yuan X, Yao Z et al (2005c) Nucleolar localization of non-structural protein 3b, a protein specifically encoded by the severe acute respiratory syndrome coronavirus. Virus Res 114(1–2):70–79
Yuan X, Shan Y et al (2006a) Mitochondrial location of severe acute respiratory syndrome coronavirus 3b protein. Mol Cells 21(2):186–191
Yuan X, Wu J et al (2006b) SARS coronavirus 7a protein blocks cell cycle progression at G0/G1 phase via the cyclin D3/pRb pathway. Virology 346(1):74–85
Yuan X, Yao Z et al (2007) G1 phase cell cycle arrest induced by SARS-CoV 3a protein via the cyclin D3/pRb pathway. Am J Respir Cell Mol Biol 37(1):9–19
Zeng R, Yang RF et al (2004) Characterization of the 3a protein of SARS-associated coronavirus in infected vero E6 cells and SARS patients. J Mol Biol 341(1):271–279
Zhao J, Falcon A et al (2009) Severe acute respiratory syndrome coronavirus protein 6 is required for optimal replication. J Virol 83(5):2368–2373
Zhong X, Yang H et al (2005) B-cell responses in patients who have recovered from severe acute respiratory syndrome target a dominant site in the S2 domain of the surface spike glycoprotein. J Virol 79(6):3401–3408
Zhong X, Guo Z et al (2006) Amino terminus of the SARS coronavirus protein 3a elicits strong, potentially protective humoral responses in infected patients. J Gen Virol 87(2):369–373
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2010 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Schaecher, S.R., Pekosz, A. (2010). SARS Coronavirus Accessory Gene Expression and Function. In: Lal, S. (eds) Molecular Biology of the SARS-Coronavirus. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-03683-5_10
Download citation
DOI: https://doi.org/10.1007/978-3-642-03683-5_10
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-03682-8
Online ISBN: 978-3-642-03683-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)